* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download EGFR_Instructor
Gene regulatory network wikipedia , lookup
Index of biochemistry articles wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Molecular evolution wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
List of types of proteins wikipedia , lookup
Mitogen-activated protein kinase wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Biochemical cascade wikipedia , lookup
Signal transduction wikipedia , lookup
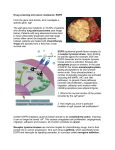
Drug screening and cancer resistance: EGFR From the gene card activity, let’s investigate a specific gene: egfr The egfr gene has mutations in 18-20% of patients who develop lung adenocarcinoma (see image below). Patients with lung adenocarcinomas tend to have unfavorable prognoses and high stage tumors often cannot be surgically removed. Instead, patients are treated with chemotherapy drugs and typically have a less than 2-year survival rate. EGFR (epidermal growth factor receptor) is a receptor tyrosine kinase. Upon binding its specific ligand (for example, EGF) the EGFR dimerizes and its receptor tyrosine kinase activity is activated. Kinases add phosphate groups to proteins. In the case of EGFR, this kinase autophosphorylates, adding phosphates to its own tyrosine amino acids. Once phosphorylated, a number of signaling cascades are activated (including the MAPK, AKT and JNK pathways). In general, these pathways promote cell proliferation, angiogenesis, migration, adhesion and invasion while inhibiting apoptosis (see adjacent figure). 1. What is the normal function of the protein encoded by the egfr gene? Encodes the EGFR membrane protein that binds extracellular ligands (ex. EGF) and activates (via phosphorylation) pathways that promote cell proliferation, angiogenesis, migration, adhesion and invasion. 2. How might you prove a particular mutation in egfr caused cell proliferation? Mutate the egfr gene in a cell line and observe the cells for hallmarks of cancer. Certain EGFR mutations cause its kinase domain to be constitutively active, meaning it can no longer be turned “off”. This causes unregulated cell proliferation, angiogenesis, migration, adhesion and invasion, all of which contribute to cancer. Receptor tyrosine kinases are common targets for chemotherapy drugs, due to their pivotal role in cancer progression. One such drug is gefitinib, which specifically binds EGFR and interrupts its signaling cascades. In a process called oncogene addiction, cancer cells tend to depend upon a single oncogenic pathway for proliferation and survival (in this case, EGFR and its downstream cascades). When this oncogenic pathway is disrupted, it leads to cancer cell death. For this reason, gefitinib is only effective on cancers with overactive/mutated EGFR. These mutations therefore render EGFR susceptible to gefitinib treatment. Gefitinib specifically inhibits the ATP binding site of the EGFP kinase domain, preventing autophosphorylation and the activation of downstream pathways. Let’s observe a lung adenocarcinoma cell line, in which various egfr mutations have been made… (image analysis activity here: 26 available alleles for EGFR). Now we have a cell culture system in which we can test chemotherapy drugs. Chemotherapy drugs with the least side effects tend to be targeted therapies, which inhibit a particular cancer-promoting pathway (like receptor tyrosine kinases). 3. View the figures below (PDB: 4WRG, 4WKQ) of EGFR wild-type and bound to gefitinib (show with black sticks). To what part of the protein does gefitinib bind? It binds in the ATP binding site Unfortunately, there are also EGFR mutations that prevent gefitinib from binding and inhibiting the kinase’s ATP binding site (see PDB 4I1Z, 4I22, 3UG2). These EGFR mutants render their cancer cells resistant to the gefitinib chemotherapy. Sometimes, when patients are treated with gefitinib, their EGFR already contains these resistant mutations. These people are called primary non-responders. In other cases, gefitinib is very effective when treatment begins, but becomes ineffective when a patient’s EGFR gains a resistant mutation. These patients have acquired resistance. 4. Where would you predict resistance mutations typically arise on the EGFR protein? How would this affect the natural function of the protein? (Interpret the figure below) Resistance mutations would affect the ATP binding site, decreasing the affinity for ATP. 5. Do the structures below confirm your hypothesis from question 4? The images below show EGFR co-crystallized with gefitinib (black sticks), the first is the “wild-type” (unmutated) structure, two structures with a susceptibility mutation (L858R and G719S, dark green sticks) and a resistance mutation (T790M, red sticks). The fourth image is a zoom in on gefitinib (black sticks) and the wild-type (light green) residues compared to the susceptibility (dark green) and resistance (red) mutations. Yes or no. Resistance mutations are found in the ATP binding domain. EFRG wild type (PDB:4wrg) EFRG + gefitinib (PDB:4wkq) EFRG (L858R/T790M)+ gefitinib (PDB:4i22) Thr 790 Ser Met 719 Gly Arg 858 EFRG (G719S/T790M)+ gefitinib (PDB:3ug2) Leu 6. When a patient seeks treatment for lung adenocarcinoma, when do you think an oncologist should prescribe gefitinib? When should gefitinib not be prescribed (use the table below)? How would the oncologist acquire this information? Sensitive/responders Exon 19 deletions Exon 21 L858R single mutation Exon 18 G719C single mutation Exon 21 L861Q single mutation Resistance/Nonresponders Exon 20 insertion Exon 20, S768I single mutation Exon 20, T790M single mutation Should be prescribed to the sensitive/responders above, should not be prescribed to the resistant/nonresponders above. This information would be acquired by sequencing parts of the patient’s genome (specifically, exons 18-21 of egfr). 7. Use the figure below to determine how common it is for a patient to be a primary nonresponder with the mutation S768I in exon 20. 0.55% of non-responders have this mutation. 8. What might be done to combat resistance mutations? Combine multiple drug treatments; don’t use any one for an extended amount of time. 9. Write a paragraph that explains the relationship between the following terms: egfr gene, EGFR protein, gefitinib, receptor tyrosine kinase, cancer and oncogene addiction. The egfr gene encodes the protein EGFR, which is a receptor tyrosine kinase. In certain cancers, EGFR has a mutation that causes it to be constitutively active, causing unregulated cell proliferation, angiogenesis, migration, adhesion and invasion and therefore cancer. The cancer cells depend upon this pathway in a process called oncogene addiction, therefore, disrupting this pathway by targeting EGFR with gefitinib kills only cancer cells. References: Cortot, A. B., and P. Janne A. "Molecular Mechanisms of Resistance in Epidermal Growth Factor Receptor-mutant Lung Adenocarcinomas." European Respiratory Review 23.133 (2014): 356-66. PyMol Molecular Viewer. Palo Alto, CA: DeLano Scientific, n.d. Computer software. RCSB Protein Databank "RCSB Protein Data Bank - RCSB PDB." RCSB Protein Data Bank - RCSB PDB. http://www.rcsb.org/pdb/home/home.do. Web. 20 Sept. 2016. Siegelin, Markus D., and Alain Borczuk C. "Epidermal Growth Factor Receptor Mutations in Lung Adenocarcinoma." Lab Invest Laboratory Investigation 94.2 (2013): 129-37. Yang, James Chih-Hsin, Yi-Long Wu, Valorie Chan, Johan Kurnianda, Kazuhiko Nakagawa, Nagahiro Saijo, Masahiro Fukuoka, Gael Mcwalter, Rose Mccormack, and Tony Mok S.k. "Epidermal Growth Factor Receptor Mutation Analysis in Previously Unanalyzed Histology Samples and Cytology Samples from the Phase III Iressa Pan-ASia Study (IPASS)." Lung Cancer 83.2 (2014): 174-81.