* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Chapter 23 – Cancer Genetics
Genomic imprinting wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Minimal genome wikipedia , lookup
History of genetic engineering wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Gene expression profiling wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
X-inactivation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Point mutation wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Designer baby wikipedia , lookup
Microevolution wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Genome (book) wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
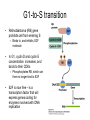
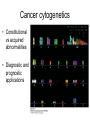
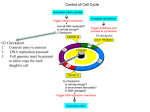
Chapter 23 – Cancer Genetics Tumor • Mass of abnormally dividing cells – Normal cells exhibit contact inhibition in culture • Benign – Usually well-defined borders; unable to metastasize • Malignant – Has ability to metastasize – “cancer” Knudson’s multistep model of cancer • Retinoblastoma – Cancer of retina – Sporadic • Unilateral; adults affected • One cell needs to accumulate mutations in both alleles – Familial • Bilateral; seen in children • One mutated allele is inherited; seen in every cell • Only one additional mutation is required Multistep model of cancer cont Clonal evolution • One cell acquires a mutation which is passed to all daughter cells • Over time, additional mutations accumulate • Genes that are involved with DNA repair or proper chromosome segregation are involved with cancer Oncogenes • Overstimulate cell division • Normal form of the gene is a proto-oncogene – Produces growthstimulating factors – Mutates into an oncogene, which hyperstimulates the cell • Dominant – Only one allele needs to be mutated to show effects Viruses associated with cancer • Can carry host proto-oncogenes – Can mutate into an oncogene which is then introduced into the host • Can interrupt normal proto-oncogene sequence when viral genome is inserted – retroviruses • Proto-oncogene may become overexpressed if placed near a promotor or enhancer Tumor suppressor genes • Inhibit cell division • Recessive – Both alleles must be mutated; often one is inherited Other gene effects • Loss of heterozygousity – Normal allele is lost due to deletion • Haploinsufficiency – A heterozygote for recessive genes has half the normal amount of gene product – Due to dosage ratios, a heterozygote may be affected with some type of phenotypic change Cell cycle control • 3 main checkpoints in cell cycle – G1-to-S – G2-to-M – Spindle assembly • Cyclin-dependent kinases (CdKs) – Enzymes that activate/inactivate other proteins by adding phosphate groups to them – Only functional when associated with a cyclin protein • Concentration of cyclins change throughout cell cycle; CDK concentration remains constant • Cyclin type determines which proteins will be phosphorylated G1-to-S transition • Retinoblastoma (RB) gene prohibits cell from entering S – Binds to, and inhibits, E2F molecule • In G1, cyclin D and cyclin E concentration increases, and binds to their CDKs – Phosphorylates RB, which can then no longer bind to E2F • E2F is now free – is a transcription factor that will express genes coding for enzymes involved with DNA replication G2-to-M transition • Mitosis promoting factor = cyclin B + CdK • Levels of cyclin B are low in G1, increases until critical level is reached near end of G2 • Phosphorylation of certain proteins cause: – Nuclear envelope breakdown, chromosome condensation, spindle formation • MPF destroys cyclin – causes cell to exit mitosis – Negative feedback – Without cyclin, low level of MPF causes return to Interphase Spindle-assembly checkpoint • Anaphase is not entered until all chromosomes are properly aligned – If not, cyclin B destruction is blocked, MPF remains active, and cell is stuck in mitosis Genes in cancer • DNA repair genes – Either increase rate of errors, and/or decrease repair of errors • Telomerase regulation – Inappropriate expression of telomerase • Vascularization – Growth factors stimulate angiogenesis Chromosomes in cancer • Translocations and inversions can create fusion proteins • CML t(9;22) – #22 has BCR gene; #9 has cABL (proto-oncogene) – Translocation creates a small #22 (Philadelphia chromosome) and relocated BCR to #9 – BCR-ABL creates fusion protein – more active than normal ABL gene • Unregulated, overactive cell division Chromosomes in cancer cont • Translocations and inversions can place a gene under new regulatory control • Burkitt lymphoma t 8 (cMYC) and 2, 14, or 22 (contain immunoglobin genes) • cMYC now under transcriptional control of immunoglobin genes • Becomes expressed in B cells; results in overproliferation Cancer cytogenetics • Constitutional vs acquired abnormalities • Diagnostic and prognostic applications