* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Proliferation of cells with HIV integrated into cancer genes
Therapeutic gene modulation wikipedia , lookup
History of genetic engineering wikipedia , lookup
Oncogenomics wikipedia , lookup
Metagenomics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genomic imprinting wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Gene expression programming wikipedia , lookup
Microevolution wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Designer baby wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Y chromosome wikipedia , lookup
Genome (book) wikipedia , lookup
Neocentromere wikipedia , lookup
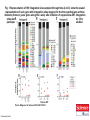
Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection by Thor A. Wagner, Sherry McLaughlin, Kavita Garg, Charles Y. K. Cheung, Brendan B. Larsen, Sheila Styrchak, Hannah C. Huang, Paul T. Edlefsen, James I. Mullins, and Lisa M. Frenkel Science Volume 345(6196):570-573 August 1, 2014 Published by AAAS Fig. 1 Representation of HIV integration sites sampled through time.(A to C) show the scaled representation of each gene with integration sites mapped for the three participants at three intervals (times in years given along the x axis) after initiation of suppressive ART. Integration sites were detected in all chromosomes of all participants, except for chromosome 18 in participant B1, and chromosome 20 and the Y chromosome in L1, the only male studied. Thor A. Wagner et al. Science 2014;345:570-573 Published by AAAS Fig. 2 Phylogenetic relationships between HIV-1 env (C2V5 region) genes sampled from participant L1 through time.A neighbor-joining tree was generated using viral gene sequences derived from PBMC DNA from participant L1 by single-genome sequencing, including from this (with integration sites determined) and a previous study (8). Thor A. Wagner et al. Science 2014;345:570-573 Published by AAAS Fig. 3 HIV-1 integration sites in chromosome 6, including the BACH2 locus.(B) (bottom) shows the integration sites mapped in the human chromosome 6 in this (SCRI, Seattle Children’s Research Institute) and other studies of individuals sampled while on suppressive ART, including from Han et al. Thor A. Wagner et al. Science 2014;345:570-573 Published by AAAS