* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download StemBase
Genome (book) wikipedia , lookup
Minimal genome wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
History of genetic engineering wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Oncogenomics wikipedia , lookup
Gene expression profiling wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
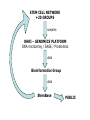
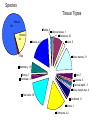
StemBase Stem Cell Network Microarray Course, Unit 6 June 2007 Sections • • • • Introduction to StemBase Using StemBase Introduction to Biomarker server Using the Biomarker server Introduction to StemBase http://www.stembase.ca The Stem Cell Genomics Project • Objective: acquire a complete understanding of the genetic factors that: – specify stem cell identity and function; and – regulate commitment and differentiation • Rationale: – Stem cells play an essential role in the human body as they provide the starting material for every organ and tissue – Knowledge of regulatory genes acting in and on stem cells is necessary to exploit their full therapeutic potential STEM CELL NETWORK +20 GROUPS samples OHRI – GENOMICS PLATFORM DNA microarray / SAGE / Proteomics data Bioinformatics Group data StemBase PUBLIC SCN Sample Contributors • • • • • • • • • • • Jane Aubin Mick Bhatia John Dick Connie Eaves Jacques Galipeau Alain Garnier Marina Gertsentein John Hassell Keith Humphries Norman Iscove Michael McBurney • • • • • • • • • • Lynn Megeney James Piret Derrick Rancourt Janet Rossant Michael Rudnicki Luc Sabourin JP Tremblay T. Michael Underhill Valerie Wallace Peter Zandstra StemBase Database of gene expression data in mouse and human stem cells Study genes important for stem cell function Affymetrix DNA microarray data (225 samples) and SAGE (6 samples) Perez-Iratxeta, C., G. Palidwor, C.J. Porter, N.A. Sanche, M.R. Huska, B.P. Suomela, E.M. Muro, P. Krzyzanowski, E. Hughes, P.A. Campbell, M.A. Rudnicki and M.A. Andrade (2005) Study of stem cell function using microarray experiments. FEBS Letters. 579, 1795-1801. Species Tissue Types Mouse 184 Retina, 3 Human 44 Muscle, 42 Adipose tissue, 1 Blastocyst, 20 Blood, 5 3 Rat Bone marrow, 31 Mammary, 14 Kidney, 1 Fibroblast, 4 Brain, 7 Calvaria, 3 Cervical epith., 3 Ciliary margin eye, 4 Fetal cells, 38 Cordblood, 11 Dermis, 1 Embryonic, 42 Genes expressed in: GO 80%100% of all samples 0%20% of all samples Pvalue GO as name GO:0030529 1.83E-66 ribonucleoprotein complex GO:0003723 1.40E-42 RNA binding GO:0005739 9.43E-31 mitochondrion GO:0006412 3.03E-28 protein biosynthesis GO:0044237 4.31E-27 cellular metabolism GO:0008380 1.79E-22 RNA splicing GO:0045184 3.50E-22 establishment of protein localization GO:0016526 3.40E-11 G-protein coupled receptor activity, unknown ligand GO:0005246 0.00362 calcium channel regulator activity GO:0007416 0.0134 synaptogenesis GO:0016358 0.0204 dendrite morphogenesis GO:0007076 0.0348 mitotic chromosome condensation Mouse / MOE430 V6.5 R1 D4A Embyoid bodies C2D C2A C2E J1 Embyoid bodies D4E Myoblasts Cancer Cancer R1 serum64 R1 serum6999 D4D Embyoid bodies Cancer Dim2 Neurospheres Retinal first passage Dermis Adipose Retinal primary Mammospheres Osteoblasts Neural Bone marrow Bone marrow Myospheres Dim1 Bone marrow Mammospheres undifferenciated Muscle Bone marrow Human / HGU133 Cord blood Bone marrow Dim2 M-O7e Smad7 Cord blood M-O7e Peripheral M-O7e Fetal Peripheral Hela I6 Cord blood Hela Bone marrow Kidney Retinal primary Myoblasts Retinal first passage Myoblasts differentiated Dim1 I6 Public web server http://www.scgp.ca:8080/StemBase/ Marker detection Use microarray data to identify probe sets that can act as markers. What and how they mark is a separate issue. Krzyzanowski and Andrade (2007) Identification of novel stem cell markers using gap analysis of gene expression data. Submitted. Method Overview • Identification – Find probe sets which appear to divide samples into two groups binary classifications Method Overview • Identification – Find probe sets which appear to divide samples into two groups binary classifications • Cluster expansion – Identify clusters of probe sets which support binary classifications • Receiver Operator Characteristic (ROC) curves are used to generate clusters of probe sets with expression patterns which can reproduce each binary classification. 1402235_at “Pattern X” 1101100101101110 Proposed markers for “Pattern X” Probe set Score 1402235_at 1.00000 1409748_at 1.00000 1430293_x_at 0.99302 … 1427392_a_at 0.90021 embryonic osteoblast fibroblast P19 spheres hematopoietic Five fold enrichment on 71 stem cell markers. From 71 in about 30,000 genes to 49 in 4,449 genes http://www.ogic.ca/projects/markerserver/enter.php Pointers • StemBase. http://www.stembase.ca • Biomarker server. http://www.ogic.ca/projects/markerserver • Thanks for your attention!