* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Bioinformatics - University of Maine System
Genomic library wikipedia , lookup
Human Genome Project wikipedia , lookup
Genome editing wikipedia , lookup
Helitron (biology) wikipedia , lookup
Pathogenomics wikipedia , lookup
Metagenomics wikipedia , lookup
Microevolution wikipedia , lookup
DNA barcoding wikipedia , lookup
Species distribution wikipedia , lookup
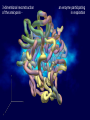
Bioinformatics GIS Applications Anatoly Petrov Bioinformatics (in a strict sense) a branch of science dealing with storage, retrieval and analysis or prediction of the composition or the structure of biomolecules (sequence analysis) - nucleic acids (DNA, RNA) - genomics - proteins - proteomics (in a wider sense) the intersection of biology and computer science (eg., computational biogeography) Bioinformatics Institute (VBI) sponsored a major conference focused on the interface between GIS and bioinformatics: “GIS Applications to Bioinformatics” (May 16–17, 2001, Blacksburg, Virginia). A Gene Map of the Human Genome The Human Transcript Map Chromosome X 3-dimentional reconstruction of the amicyanin - an enzyme participating in respiration Chromosome structure Nucleosome Chromosomal DNA is packaged into a compact structure with the help of specialized proteins called histones. The fundamental packing unit is known as a nucleosome. Sequence features that appear to be spatially disconnected according to a linear representation of a genome, may actually be close neighbors due to the folding of DNA into a 3-dimensional molecule. GenoSIS - Genome Spatial Information System (ESRI ArcGIS – visualization tool + Oracle Spatial – object-relational database) Applications: - thematic mapping and visualization exploratory spatial data analysis (ESDA) Set of questions for the ESDA · Where do we find consensus sequence elements (CSEs)? How many elements are there at that genomic region? · Is there regularity in their distribution? What is the nature of that regularity? Why should the spatial distributional pattern exhibit regularity? · Are CSEs found throughout the genome? What are the limits to where they are found? Why do those limits constrain its distribution? · Are there regulatory elements spatially associated with a gene with a particular molecular function? Do these regulatory elements and genes usually occur together in the same places? Why should they be spatially associated? · Has a particular gene always been there? In which organism did it first emerge or become obvious? How has it changed spatially (through evolutionary time)? · What factors have influenced its duplication or deletion in the genome? What factors have constrained its spread? Using GIS for thematic genome mapping Application of ArcView for genome mapping and spatial analysis Biogeography Prediction (reconstruction) of species distribution - In the past (paleobiogeography and paleoclimatology; ex., NOAA’s Paleoclimatology Program) - At present (eg., Environment Australia’s Species Mapper) - In future (eg., some of the Lifemapper products) Methods There are two main datasets that are fundamental in obtaining good prediction on a species distribution map: species occurrence data and environmental information. Algorithms GARP, environmental classification method) envelope (BIOCLIM), e-ball, “image” (Bayesian Habitat Digitizer Extension (HDE) to ArcView uses a hierarchical classification scheme to delineate habitats by visually interpreting georeferenced images such as aerial photographs, satellite images, and side scan sonar. HDE allows users to create custom classification schemes and rapidly delineate and attribute polygons using simple menus. Deducing potential species distribution using BIOCLIM Environment Australia’s Species Mapper Query database to retrieve records of species locations. For each species location, interpolate values of essential climatic variables. Calculate the climatic envelope bounding all the species records. At the resolution specified, identify all other sites in the landscape that fall within the climatic envelope. Plot the sites identified on a base map. Deliver the map to the user. Lifemapper 1. The species occurrence data is gathered from a number of biological collections housed at several museums and herbaria worldwide. Those institutions have their specimen databases linked and integrated through The Species Analyst project. 2. The environmental information is represented as a set of geographic layers. Each layer displays one particular environmental parameter, such as temperature, rainfall, land use, elevation, among others. 3. Using data from those two datasets, GARP tries to find nonrandom correlations between species occurrence data and the values of the environmental parameters where the species occur or do not occur. G Genetic A Algorithm for R Ruleset P Production Paleobiogeography 3-D Flythrough animation Holocene Evolution of the Southern Washington and Northern Oregon Shelf and Coast. NOAA’s Paleoclimatology Program Pollen Viewer THE END