* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download ppt presentation
Gel electrophoresis of nucleic acids wikipedia , lookup
Synthetic biology wikipedia , lookup
Molecular cloning wikipedia , lookup
Comparative genomic hybridization wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Molecular ecology wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Personalized medicine wikipedia , lookup
Non-coding DNA wikipedia , lookup
Community fingerprinting wikipedia , lookup
Genome evolution wikipedia , lookup
Molecular evolution wikipedia , lookup
DNA sequencing wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
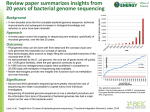
Genomics Gene expression • Genome maping • Genome sequencing • Genome annotations Structural genomics Nucleus DNA (Genome) pre-mRNA Cytoplasm • DNA arrays and chips • (semi) qRT-PCR • Northern blot + hybrid. • Transkriptional fusions mRNA mRNA (Transcriptome) Proteins (Proteome) Metabolites (Metabolome) • 2D electrophoresis Mass spectrometry Protein sequencing • Translational fusional • Immunodetection • Enzyme activities • Chromatography • Mass spectrometry • NMR Functional genomics History of genomes sequencing • 1977 bacteriophage øX174 (5386bp, 11 genes) • 1981 mitochondrial genome (16,568bp; 13 prots; 2 rRNAs; 22 tRNAs • 1986 chloroplast genome (120,000-200,000bp) • 1992 Saccharomyces chromosome III (315kb; 182 ORFs) • 1995 Haemophilus influenzae (1.8Mb • 1996 Saccharomyces whole genome (12.1Mb; over 600 people 100 laboratories) • 1997 E. coli (4.6Mb; 4200 proteins) • 1998 Caenorhabditis elegans (97 Mb; 19,000 genů) • 2000 Arabidopsis thaliana (115Mb, 25-30,000 genů) • 2001 mouse (1 year!) • 2001 Homo sapiens (2 projekty) • 2005 Pan, rice • 2006 Populus Technological improvements DNA sequencing – principle (Sanger’s method) Polymeration from primer in the presence of low concentration of terminator (dideoxy) ddNTP primer Random termination on all positions with occurance of the nucleotide Original arrangement sequence - RI labelled primer - 4 separated reactions - with individual ddNTP - ddNTP:dNTP (cca 1:20 – (100)) - PAGE separation A T C G C T G G A T C T A G C Separation by size Automated sequencing with fluorescence-labelled ddNTP • Every ddNTP labelled with different fluorescent • dye – all together in one reaction Separation by size in capillary – fluorescence detection Genom sequencing is more than sequencing of DNA • 1 sequencing reaction 300 – 800 bp • Typical genom hunderts of millions to billions bp How to manage? Strategies of genome sequencing • Classical strategy (Map-Based Assembly): - minimal quantity of DNA sequencing – sorting of big DNA fragments, successive reading (human genome sequencing – original strategy) - scaffold for genome sequence assemble - time consuming • Whole genome shotgun (WGS) – random (7-9x redundant) sequencing – sorting of sequence data (Haemophilus) - problems with repetitive DNA • Combination – „hierarchical shotgun“, „chromosome shotgun“ Hierarchical shotgun sequencing Whole-genome shotgun sequencing Production of overlapping clones (e.g. BACs, YACs) and construction of physical map Shearing of DNA and sequencing of subclones Assembly Green (2001) Nature Reviews Genetics 2: 573-583 Hierarchical shotgun sequencing First step: library of big DNA inserts (= genome fragments) • • • • phage (l) vectors: 30 kb cosmids: 50 kb BACs (bacterial artificial chromosomes): 100-300 kb YACs (yeast artificial chromosomes): cca 0.5-1Mb Physical „BAC“ map of genome • Arrangement (position, orientation) of individual BAC in the genome • Fundamental for classical sequencing • Very usefull for assembly of „shotgun“ sequences How to make the map from BACs with unknown sequence? Map construction - BAC fingerprinting Sequencing of DNA ends Restriction sites - 10-20x more bp in BACs than in the genome for map construction (Arabidopsis – 20 000, rice - 70 000) BAC fingerprinting ANIMATION of HIERARCHICAL SHOTGUN: http://www.weedtowonder.org/sequencing.html Minimum tiling path = the lowest possible set of BACs covering the whole sequence physical map arrangement and mapping and clone selection - by restriction fragment analysis - using terminal sequences and hybridization - by hybridization with markers with known position in genetic map Shotgun sequencing BAC/chromosome/whole genome random cleavage + direct sequencing (NGS) Cosmids (40 Kbp): sequencing of clone ends (known distance between) ~500 bp ~500 bp Genome (chromosome, BAC...) assembly 1. Looking for overlaps in primary sequences 2. Assembly to contigs to get short consensus sequences 3. Assebly to supercontigs using the information of sequence pairs (ends + distance) 4. Complete consensus sequence ..ACGATTACAATAGGTT.. Repetitive sequences and contig assembly repetition ? ? Repetitions are serious problem in assembly, if they are conserved and longer than sequencing run Use of markers for whole genome assembly (STS – sequence tagged sites = short sequences with known position on chromosoms) Supecontigs with scaffold (BAC-end sequences with known distance) Filling of gaps: shorter clones are better X - optimal – libraries with different insert sizes (2, 10, a 50 kbp) - sequencing the linker clone = filling the gap What to do with the genome sequence? To annotate! • Searching for genes: – – – – Automatic prediction of coding seq. Prediction of introns/exons Prediction according to related seq. Confirmation by cDNA and EST • Prediction of function – from experimentally characterized homologues Fragment of GenBank BAC clone annotation Graphical interface of BAC annotation Large genomes alternative strategies of sequencing: - isolation of individual chromosomes e.g. wheat – allows assembly of homeologous chromosomes (allohexaploid) - shotgun sequencing of non-methylated DNA (maize) - sequencing of ESTs (potato) Expressed Sequence Tags (ESTs) -short sequenced regions of cDNA (300-600 nt) -usually gene fragments (primarilly originate from mRNA) -highly redundant, but also incomplete! -problems: - no regulatory sequences (promotors, introns,...) - only transcripts of certain genes Expressed Sequence Tags (ESTs) Preparation of EST library - mRNA - RT with oligoT primer cDNA -cleavage of RNA from heteroduplex RNAseH - 2nd strand cDNA synthesis - cleavage with restriction endonuclease - adaptor ligation cloning sequencing Assembly of EST contigs - Unigenes Next generation sequencing - faster and cheaper!!! - parallel sequencing of high numbers of sequences! - no handling with individual sequences! Examples of recently developed or developing technologies: 454 sequencing – pyrosequencing (Roche) - complementary strand synthesis Illumina – sequencing by synthesis - complementary strand synthesis SOLiD - Sequencing by Oligonucleotide Ligation and Detection - ligation of labelled oligonucleotides Oxford nanopore technology - exonuclease degradation, el. current changes detection NGS – comparison of basic parameters Method Single-molecule real-time sequencing (Pacific Bio) Ion Sequencing by semiconductor Pyrosequencing synthesis (Ion Torrent (454) (Illumina) sequencing) Read length 5.000-10.000 (30.000) bp up to 400 bp Reads per run 50.000 Cost per 1 million bases (in US$) $0.33-$1.00 700 bp Sequencing by ligation (SOLiD sequencing) Chain termination (Sanger sequencing) 50 to 300 bp 50+50 bp up to 80 million 1 million up to 3 billion 1.2 to 1.4 billion N/A $1 $0.05 to $0.15 $0.13 $10 400 to 900 bp $2400 http://en.wikipedia.org/wiki/DNA_sequencing 454 technology - pyrosequencing up to 1 mil reads (lenght 700 - 1000 bp) one day (23 hour procedure) = 500-800 Mbp 454 technology - pyrosequencing 454 technology 454 technology Illumina – sequencing by synthesis (Solexa) Illumina – seqencing by synthesis (Solexa) Illumina – seqencing by synthesis (Solexa) Illumina – seqencing by synthesis (Solexa) SOLiD™ System (Applied Biosystems) 2 Base Encoding Sequencing by Oligonucleotide Ligation and Detection - reads up to 75 b - 20-30 Gb for a day! - high accuracy up to 99,99 % - initial step – clonal multiplication (similar to 454) http://appliedbiosystems.cnpg.com/Video/flatFiles/699/index.aspx SOLiD™ System Mix of 1024 octamers (number of variations NNN = 64) x 16 known dinucleotides Z = nucleotides universally pairing with any nucleotide (prolongation) – cleaved out after ligation labelling: 4 fluorescent dyes – each for 256 octamers (with just 4 known middle dinucleotides) - 5 independent reactions = each 10 – 15 times repeated ligations of labelled octamers starting from a primer with shifted end Knowledge of the first nucleotide allows translation of color sequence to nucleotide sequence AAT G CA GGCATG CCGTAC } alternative translation with different 1st nucleotide Oxford nanopore technologies – direct sequencing http://www.nanoporetech.com/sequences of one DNA strand - protein nanopore in membrane (alpha-hemolysin) - covalently bound exonuclease - monitoring specific decrease in current (metC!)