* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download slides
Cell culture wikipedia , lookup
Extracellular matrix wikipedia , lookup
Cell growth wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
Cytokinesis wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
Signal transduction wikipedia , lookup
Lipopolysaccharide wikipedia , lookup
Programmed cell death wikipedia , lookup
List of types of proteins wikipedia , lookup
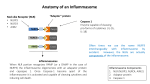
Epithelial Cells and Their Role in Immunity Dr. Irving Coy Allen Virginia Tech VA-MD Regional College of Veterinary Medicine Department of Biomedical Sciences and Pathobiology Mucosal Immunology: An Overview Animation Developed by Tom MacDonald http://www.nature.com/ni/multimedia/mucosal/index.html AB/PAS Staining Goblet Cell and Mucus Staining Mock Mock AOM/DSS AOM/DSS Three Classes of Pattern-Recognition Receptors Modified From: Osamu Takeuchi and Shizuo Akira; Immunological Reviews (2009) TLR RLR TLR-7 TLR-3 MyD88 TRIF RIG-I NLR MDA5 NLR Inflammasome MAVS cleavage IRF7 IRF3 NF-κB Cytokine Genes Type-I IFN Genes pro-IL-1β pro-IL-18 IL-1β IL-18 NLRs in Intestinal Homeostasis NLR SPECIFICITY Inflammasome Forming NLRs NLRP3 K+ ASC Speck IFNγR Cytosolic Gram Negative Bacteria priming STAT1 ROS mtDNA Ca2+ IFNαR TLR3 Toxins TLR4 DAMPs IFN-β IFN-γ LPS polyI:C TNF TNFR ATP Crystals LPS Hypothetical Non-Canonical Inflammasome Caspase-11 GBP5 Caspase-1 p10 pro-Caspase-1 NLRP3 Oligomer pro-IL-1β pro-IL-18 p20 IL-1β IL-18 Pyroptosis NLRP6 Menno van Lookeren Campagne & Vishva M. Dixit Nature 474, 42–43 (02 June 2011) doi:10.1038/474042a Pyroptosis Nat Rev Microbiol. 2009 February ; 7(2): 99–109. Pathogen Modulation of Inflammasome Function Inhibitory NLRs Non-canonical NF-κB Signaling Canonical NF-κB Signaling TLR MyD88 TRAF3 TRAF6 NIK NLRC3 Cytosol IKK-γ NF-κB CD40 NLRP12 IRAK1 NLRX1 Extracellular IKK-α/α IKK-α/β IκB p50 p65 p50 p65 NF-κB Binding Motif p52 Rel B Inflammation Migration Differentiation Invasiveness Survival p100 Rel B p52 Rel B NF-κB Binding Motif Nucleus Utilizing Mitochondria as Scaffolding RNA NLRX1 RIG-I NLRX1 MAVS RIG-I NLRX1 MAVS UQCRC2 TBK1 NLRX1 TUFM ATG16L1 ATG5 ATG12 Mitochondria IRF IFN-I IL-6 Autophagy ROS Computational modeling should prove highly useful to elucidate the complex interplay between immunity, metabolism and the microbiota, and provide insight on pharmacokinetic and pharmacodynamic regulation of new IBD therapies Predicting IBD prognosis is patient-specific, time sensitive and often elusive, yet crucial for deciding effective treatment and disease control. Mathematical and computational modeling offers a novel perspective for identifying molecular targets aimed at the development of more efficacious and safer personalized interventions for IBD. Publically available microarray studies offer robust datasets for calibrating, or fitting, mathematical equations to observed biological phenomenon. Computational Modeling: PRRs and Epithelial Cell Pathobiology Reconciling conflicting genomic results, integrating transcriptomics, proteomics, flow cytometry and histology data with specific clinical outcomes in patients with wellcharacterized gene variants through bioinformatics and computational modeling approaches would provide an invaluable assessment of TLR and NLR functionality among heterogeneous populations of IBD patients. Computational modeling could be used to investigate cell specificity of NLRs and the role of NLRs in sensing dysbiosis in addition to mechanisms underlying modulation of T cell differentiation by dysregulated NLR signaling. Modeling: Structure-Based Virtual Screening (SBVS) Questions? Irving Coy Allen [email protected] http://upload.wikimedia.org/wikipedia/commons/0/0b/Burruss_Hall,_Virginia_Tech.JPG