* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Diapositiva 1 - Universidad Autónoma de San Luis Potosí
Survey
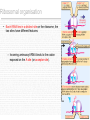
Document related concepts
Polycomb Group Proteins and Cancer wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Frameshift mutation wikipedia , lookup
Polyadenylation wikipedia , lookup
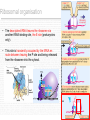
History of RNA biology wikipedia , lookup
Point mutation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Non-coding RNA wikipedia , lookup
Primary transcript wikipedia , lookup
Messenger RNA wikipedia , lookup
Genetic code wikipedia , lookup
Expanded genetic code wikipedia , lookup
Epitranscriptome wikipedia , lookup
Transcript
Prokaryote Translation CA García Sepúlveda MD PhD Laboratorio de Genómica Viral y Humana Facultad de Medicina, Universidad Autónoma de San Luis Potosí 1 Introduction • Ribosomes provide the environment for controlling the interaction between mRNA and aminoacyl-tRNA. • Behave like a small migrating factories travelling along the mRNA template engaging in rapid cycles of peptide bond synthesis. • Possess several active centers, each constructed from a particular group of proteins associated with rRNA. • The active centers require rRNA for structural and catalytic roles. • Some catalytic functions require particular proteins, but none of the activities can be reproduced by isolated proteins or groups of proteins; they function only in the context of the ribosome (associated with RNA). 2 Molecular Proportions • Each ribosome subunit has specific roles. • mRNA is associated with the small subunit (~30 bases of the mRNA). • tRNAs are quite large relative to the ribosome; they become inserted into internal sites that stretch across the subunits. • A third tRNA may remain present on the ribosome after it has been used in protein synthesis, before being recycled (prokaryotes only). • Polypeptide elongation involves reactions at just two of the ~10 codons covered by the ribosome. 3 Ribosomal organisation • Each tRNA lies in a distinct site on the ribosome, the two sites have different features: – Incoming aminoacyl-tRNA binds to the codon exposed on the A site (or acceptor site). 4 Ribosomal organisation • Each tRNA lies in a distinct site on the ribosome, the two sites have different features: – A salient peptidyl-tRNA (tRNA carrying the polypeptide) occupies the codon in the P site (or donor site). 5 Ribosomal organisation • Each tRNA lies in a distinct site on the ribosome, the two sites have different features: – The end of the tRNA that carries an amino acid is located on the large subunit. 6 Ribosomal organisation • Each tRNA lies in a distinct site on the ribosome, the two sites have different features: – The anticodon at the other end interacts with the mRNA bound by the small subunit. – So the P and A sites each extend across both ribosomal subunits. 7 Ribosomal organisation • Each tRNA lies in a distinct site on the ribosome, the two sites have different features: – Peptide bond formation occurs when the polypeptide carried by the peptidyl-tRNA is transferred to the amino acid carried by the aminoacyl-tRNA. – This reaction is catalyzed by constituents of the large subunit of the ribosome. 8 Ribosomal organisation • Transfer of the polypeptide generates the ribosome shown in step 2. • The deacylated tRNA, lacking any amino acid, lies in the P site, while a new peptidyl-tRNA has been created in the A site. • This peptidyl-tRNA is one amino acid residue longer than the peptidyl-tRNA that had been in the P site in step 1. 9 Ribosomal organisation • Then the ribosome moves one triplet along the messenger = TRANSLOCATION. 10 Ribosomal organisation • Then the ribosome moves one triplet along the messenger = TRANSLOCATION. • The movement transfers the deacylated tRNA out of the P site... 11 Ribosomal organisation • Then the ribosome moves one triplet along the messenger = TRANSLOCATION. • The movement transfers the deacylated tRNA out of the P site... • moves the peptidyl-tRNA into the P site. 12 Ribosomal organisation • Then the ribosome moves one triplet along the messenger = TRANSLOCATION. • The movement transfers the deacylated tRNA out of the P site... • moves the peptidyl-tRNA into the P site. • Exposes the next codon to be translated in the A site, ready for a new aminoacyl-tRNA to enter. 13 Ribosomal organisation • The deacylated tRNA leaves the ribosome via another tRNA-binding site, the E site (prokaryotes only). • This site is transiently occupied by the tRNA en route between leaving the P site and being released from the ribosome into the cytosol. 14 The three stages Protein synthesis divided into the three stages: 1.- Initiation 2.- Elongation 3.- Termination 15 The three stages Initiation involves the reactions that precede formation of the peptide bond between the first two amino acids of the protein. It requires the ribosome to bind to the mRNA, forming an initiation complex that contains the first aminoacyl-tRNA. This is a relatively slow step in protein synthesis, and usually determines the rate at which an mRNA is translated. 16 The three stages Elongation includes all the reactions from synthesis of the first peptide bond to addition of the last amino acid. Amino acids are added to the chain one at a time. This is the most rapid step in protein synthesis. 17 The three stages Termination encompasses the steps that are needed to release the completed polypeptide chain and dissociate the ribosome from the mRNA. 18 Accessory Factors • Different sets of accessory factors assist the ribosome at each stage. 19 Accessory Factors • Different sets of accessory factors assist the ribosome at each stage. • Energy is provided at various stages by the hydrolysis of GTP. 20 Accessory Factors • Different sets of accessory factors assist the ribosome at each stage. • Energy is provided at various stages by the hydrolysis of GTP. • Accessory factors are not covalently linked to Ribosome (proteins or RNA). 21 Initiation - Prokaryotes Initiation in bacteria needs 30S subunits and accessory factors • Initiation factors (IF in prokaryotes, eIF in eukaryotes) are proteins that associate with the small subunit of the ribosome during initiation. • Initiation of protein synthesis is not a function of intact ribosomes, but is undertaken by the separate subunits, which reassociate during the initiation reaction. • See “Ribosomal subunit cycle during protein synthesis in bacteria”. 22 Initiation – Subunit Cycle Ribosomal subunits cycle during prokaryote protein synthesis. 23 Initiation – Subunit Cycle Ribosomal subunits cycle during prokaryote protein synthesis. Bacterial ribosomes engaged in elongating a polypeptide exist as 70S. 24 Initiation – Subunit Cycle Ribosomal subunits cycle during prokaryote protein synthesis. Bacterial ribosomes engaged in elongating a polypeptide exist as 70S. At termination, they are released from the mRNA to enter a pool of free ribosomes. 25 Initiation – Subunit Cycle Ribosomal subunits cycle during prokaryote protein synthesis. Bacterial ribosomes engaged in elongating a polypeptide exist as 70S. At termination, they are released from the mRNA to enter a pool of free ribosomes. In growing bacteria, 80% of ribosomes are synthesizing proteins only 20% are free. 20% 80% 26 Initiation – Subunit Cycle Ribosomal subunits cycle during prokaryote protein synthesis. Bacterial ribosomes engaged in elongating a polypeptide exist as 70S. At termination, they are released from the mRNA to enter a pool of free ribosomes. In growing bacteria, 80% of ribosomes are synthesizing proteins only 20% are free. Ribosomes in the free pool can dissociate into separate subunits. Free 70S ribosomes are in dynamic equilibrium with 30S and 50S subunits. 27 Initiation - Prokaryotes • Initiation occurs at a special sequence on mRNA called the ribosome-binding site. • Short sequence of bases that precedes the coding region. • The ribosome-binding site is a sequence at which the small and large subunits associate on mRNA to form an intact ribosome. AGGAGG AUG 28 Initiation - Prokaryotes The reaction occurs in two steps: Recognition of mRNA occurs when a small subunit binds to form an initiation complex at the ribosome-binding site. 29 Initiation - Prokaryotes The reaction occurs in two steps: Recognition of mRNA occurs when a small subunit binds to form an initiation complex at the ribosome-binding site. Then a large subunit joins the complex to generate a complete ribosome. 30 Initiation Factors - Prokaryotes • The 30S subunit is involved in initiation but not by itself competent to undertake the reactions of binding mRNA and tRNA. • Requires additional proteins called initiation factors (IF). • These factors are found only on 30S subunits, and they are released when the 30S subunits associate with 50S subunits to generate 70S ribosomes. 31 Initiation Factors - Prokaryotes • The 30S subunit is involved in initiation but not by itself competent to undertake the reactions of binding mRNA and tRNA. • Requires additional proteins called initiation factors (IF). • These factors are found only on 30S subunits, and they are released when the 30S subunits associate with 50S subunits to generate 70S ribosomes. NOTE: • • • • This behavior distinguishes IF from the structural proteins of the ribosome. IF's are concerned solely with formation of the initiation complex. They are absent from 70S. They play no part in the stages of elongation. 32 Initiation Factors - Prokaryotes Bacteria use three initiation factors: IF-1, IF-2 and IF-3. IF-3 is needed for 30S subunits to bind to initiation sites in mRNA. 33 Initiation Factors - Prokaryotes Bacteria use three initiation factors: IF-1, IF-2 and IF-3. IF-3 is needed for 30S subunits to bind to initiation sites in mRNA. IF-2 binds a special initiator tRNA and controls its entry into the 30S P site. 34 Initiation Factors - Prokaryotes Bacteria use three initiation factors: IF-1, IF-2 and IF-3. IF-3 is needed for 30S subunits to bind to initiation sites in mRNA. IF-2 binds a special initiator tRNA and controls its entry into the 30S P site. IF-1 binds to 30S and stabilize the complete initiation complex. 35 Initiation Factors - Prokaryotes IF-3 has two functions: 1.- ANTI-ASSOCIATION FACTOR. Stabilizes free 30S subunits. Which controls the amount of free 30S subunits, preventing them from reassociating with 50S subunits. 36 Initiation Factors - Prokaryotes IF-3 has two functions: 2.- BINDING FACTOR. Controls the ability of 30S subunits to bind to mRNA. Small subunits must have IF-3 in order to form initiation complexes with mRNA. IF-3 must be released from the 30S:mRNA complex in order to enable the 50S subunit to join. On its release, IF-3 immediately recycles by finding another 30S subunit. 37 Initiation – Initiator tRNA • A special initiator tRNA starts the polypeptide chain • Synthesis of all proteins starts with the same amino acid: methionine. • The signal for initiating a polypeptide chain is a special initiation codon that marks the start of the reading frame. 38 Initiation – Initiator tRNA • Usually the initiation codon is the triplet AUG, but in bacteria, GUG or UUG are also used. • The AUG codon represents methionine and initiations. • How are they differentiated by translational machinery? 39 Initiation – Initiator tRNA • Two types of tRNA can carry an anticodon for these codons. • One tRNA is used for initiation, the other for recognizing AUG codons during elongation. • Prokaryote initiator tRNA is a special tRNA that is formylated (thus the name tRNAf). • It is linked to a Methionine residue (aminoacyl-tRNA), thus the name tRNAfMet • As Met residue is also formylated it is also abbreviated fMet-tRNAf. 40 Initiation – Initiator tRNA • Formylation is a two stage reaction. • First, tRNA is charged with Met to generate Met-tRNAf. • Then formylation blocks the free NH2 group. Formyl blockage prevents incorporation to the A site (elongation) but can be loaded into P site. 41 Initiation – Initiator tRNA • This tRNA is used only for initiation. • It recognizes the codons AUG or GUG (occasionally UUG). • The codons are not recognized equally well: the extent of initiation declines 50% when AUG is replaced by GUG. • Inititation declines by 75% when AUG is replaced by UUG. • The species responsible for recognizing AUG codons in internal locations is tRNAmMet. • This tRNA responds only to internal AUG codons. • Its methionine cannot be formylated. 42 Initiation – Initiator tRNA • So there are two differences between the initiating and elongating Met-tRNAs: the tRNA moieties themselves are different; and the amino acids differ in the state of the amino group. • The meaning of the AUG and GUG depends on their context. – When the AUG codon is used for initiation, it is read as formyl-Met; when used within the coding region, it represents Met. • The meaning of the GUG codon is even more dependent on its location. – When present as the first codon, it is read via the initiation reaction as formylMet yet when present within a gene, it is read as Val. 43 Initiation – Initiator tRNA 44 Initiation – Initiator tRNA • In an initiation complex, the small subunit alone is bound to mRNA. • The initiation codon lies within the part of the P site. • The only aminoacyl-tRNA that can become part of the initiation complex is the initiator, which has the unique property of being able to enter directly into the partial P site to recognize its codon. 45 Initiation – Initiator tRNA • When the large subunit joins the complex, the initiator fMet-tRNAf lies in the now-intact P site. • The A site is available for entry of the aminoacyl-tRNA complementary to the second codon of the gene. 46 Initiation – Initiator tRNA • Initiation prevails when an AUG (or GUG) codon lies within a ribosome-binding site, because only the initiator tRNA can enter the partial P site generated when the 30S subunit binds de novo to the mRNA. • Only the regular aminoacyl-tRNAs can enter the (complete) A site. 47 Initiation – Initiator tRNA • What features distinguish the tRNAfMet initiator and the tRNAmMet elongator? • Some of these features are needed to prevent the initiator from being used in elongation, others are necessary for it to function in initiation 48 Initiation – Initiator tRNA Formylation Both tRNA and amino acid Formylation is not strictly necessary, because nonformylated tRNAfMet can function as an initiator. Formylation improves the efficiency with which the tRNAfMet is bound by IF-2. 49 Initiation – Initiator tRNA Acceptor Stem base-pairing The bases C:A on the acceptor stem are NOT paired in tRNAfMet. The absence of this pairing makes tRNAfMet uncapable of being incorporated during elongation. Mutations that create a base pair in this position of tRNAfMet allow it to function in elongation. It is also needed for the formylation reaction. 50 Initiation – Initiator tRNA Anti-codon stem base-pairing A series of 3 G:C pairs in the stem that precedes the loop containing the anticodon is unique to tRNAfMet. Are required to allow the tRNAfMet to be inserted directly into the P site. 51 Initiation – Initiator tRNA The ability of tRNAfMet initiator to enter the ribosome is controlled by IF-2. The 30S subunit carries all the initiation factors, including IF-2, which is probably associated with the P site. IF-2 specifically forms a complex with tRNAfMet. This complex places the tRNA in the partial P site. 52 Initiation – Initiator tRNA By forming a complex specifically with tRNAfMet, IF-2 ensures that only the initiator tRNA, and none of the regular aminoacyl-tRNAs, participates in the initiation reaction. IF-2 remains part of the 30S subunit at this stage; it has a further role to play... 53 Initiation – Initiator tRNA IF-2 has a ribosome-dependent GTPase activity. It sponsors the hydrolysis of GTP in the presence of ribosomes, releasing the energy stored in the high-energy bond. The GTP is hydrolyzed when the 50S subunit joins to generate a complete 70S ribosome. 54 Initiation – Initiator tRNA In bacteria and mitochondria, the formyl residue on the initiator methionine is removed by a specific deformylase enzyme to generate a normal NH2 terminus. 50% of the proteins the methionine at the terminus is removed by an aminopeptidase, creating a new terminus from R2 (the second amino acid incorporated into the chain). When both steps are necessary, they occur sequentially. The removal reaction(s) occur rather rapidly, probably when the nascent polypeptide chain has reached a length of 15 amino acids. 55 Initiation – Shine-Dalgarno Sequences Initiation involves base pairing between mRNA and rRNA An mRNA contains many AUG triplets: how is the initiation codon recognized as providing the starting point for translation? When ribonuclease is added to the blocked initiation complex, all the regions of mRNA outside the ribosome are degraded. The protected fragments can be recovered and characterized. 56 Initiation – Shine-Dalgarno Sequences The protected initiation sequences of bacteria are ~30 bases long. Two common features: The AUG (or less often, GUG or UUG) initiation codon is always included within the protected sequence. Within 10 bases upstream of the AUG is a polypurine stretch known as the ShineDalgarno sequence: 5’-AGGAGG-3’ It is complementary to a highly conserved sequence close to the 3’ end of 16S rRNA: 3’-UCCUCC-5’ 57 Initiation – Shine-Dalgarno Sequences Start Codon AUG Shine-Dalgarno sequence (in mRNA): 5’-AGGAGG-3’ Shine-Dalgarno complementary sequence (16S rRNA): 3’-UCCUCC-5’ 58 Initiation – Shine-Dalgarno Sequences Mutations of any of these sequences impede ribosome binding. Point mutations in the Shine-Dalgarno sequence prevent mRNA translation. The sequence at the 3’ end of rRNA is conserved between prokaryotes and eukaryotes... 59 Initiation – Shine-Dalgarno Sequences However, eukaryotes have a five-base deletion in the complementary rRNA sequence. There is no apparent base pairing between eukaryotic mRNA & 18S rRNA. This is a significant difference in the mechanism of initiation. 60 Initiation – Shine-Dalgarno Sequences In bacteria, a 30S subunit binds directly to a ribosome-binding site. As a result, the initiation complex forms at a sequence surrounding the AUG initiation codon. When the mRNA is polycistronic, each coding region starts with a ribosome-binding site. 61 Initiation – Shine-Dalgarno Sequences When ribosomes attach to the first coding region, the subsequent coding regions have not yet even been transcribed. By the time the second ribosome site is available, translation is well under way through the first cistron. 62