* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Ph1
Nutriepigenomics wikipedia , lookup
Segmental Duplication on the Human Y Chromosome wikipedia , lookup
Microevolution wikipedia , lookup
History of genetic engineering wikipedia , lookup
Hybrid (biology) wikipedia , lookup
Histone acetyltransferase wikipedia , lookup
Designer baby wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
Human genome wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genome evolution wikipedia , lookup
Y chromosome wikipedia , lookup
Human–animal hybrid wikipedia , lookup
Genome (book) wikipedia , lookup
X-inactivation wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Genetically modified organism containment and escape wikipedia , lookup
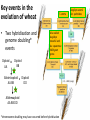
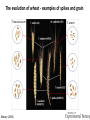
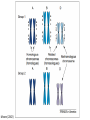
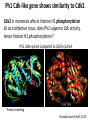
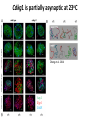
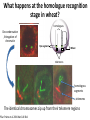
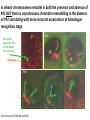
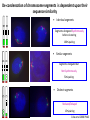
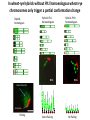
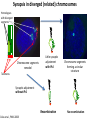
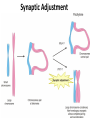
The Ph Locus and the rise of bread wheat Dr Glyn Jenkins Wheat – a plant that feeds the world Cultivated area: 215,489,485 Ha (area of UK 22,933,252Ha) Production: 670,775,485 tonnes Global productivity: 3.1 t/Ha Contributes 20% of total food calories and protein in human nutrition Wheat 20:20 – Project aim to have achieve an average yield of 20 t/Ha Yield plateau reached - so where do we go from here? Grassini et al. (2012) The origin of wheat • First cultivation of wheat (diploid and tetraploid) occurred about 10000 years ago, as part of the ‘Neolithic Revolution’ • Cultivation spread to the Near East • 9000 years ago hexaploid bread wheat made its first appearance • The main route into Europe via • Greece (8000 BP) • Balkans to the Danube (7000 BP) • Italy, France and Spain (7000 BP), • UK and Scandinavia by about 5000 BP Matsuoka (2011) Key events in the evolution of wheat • Two hybridisation and genome doubling* events Diploid x AA T. urartu Also called Aegilops tauschii and Ae. squarrosa Wild goat grass Diploid BB Allotetraploid x Diploid AA BB DD Allohexaploid AA BB DD *chromosome doubling may have occurred before hybridisation Aegilops searsii Ae. speltoides The evolution of wheat - examples of spikes and grain T. monococcum Shewry (2009) T. searsii Gupta et al. (2008) Bread Wheat Triticum aestivum ssp. aestivum 2n = 6x =42 – Wheat has 3 homoeologous chromosome sets A, B and D – Disomic inheritance preserves hybrid nature – Behaves as a diploid at meiosis – HOW? 1 Triticum monoccoccum A Triticum searsii B Triticum tauchii D 2 3 4 5 6 7 Meiosis Paired homologues align on plate 1 Diploid cell Prophase I Zygotene Pachytene Diplotene Leptotene Diakinesis Metaphase I Anaphase I Homologues segregate Telophase II 4 Haploid cells Telophase I Anaphase II Metaphase II Prophase II Sister chromatids segregate Incorrect pairing leads to unbalanced gametes and infertility How does wheat produce 4 haploid cells at the end of meiosis? Moore (2002) Pairing homoeologous • Initially it was assumed that the three diploid species whose genomes had gone to make up hexaploid wheat were strongly differentiated – How else could one explain the near absence of meiotic pairing in haploids of the hexaploid species? • 1952 – became clear that the corresponding chromosomes of the three different genomes are genetically very closely related • Riley and Chapman (1958) - discovered that homoeologous pairing is suppressed by a gene or genes on the long arm of chromosome 5B – Became known as Ph1 – N.B. – wheat contains additional Ph loci • How does Ph1 work? Sears (1976) Ph1 in Wheat • Led by Prof Graham Moore • Research - Wheat meiosis and the Ph1 locus http://www.jic.ac.uk /staff/grahammoore/index.htm Effect of Ph1 Ph1• Multivalents • Univalents Ph1+ Martinez et al. (2001) ∴ Ph1 is critical to maintaining genome stability in wheat Effect of Ph1 Wheat-rye hybrid Ph1 locus suppresses pairing between related chromosomes (homoeologous pairing) If Ph1 locus is deleted, pairing is induced between homoeologous chromosomes Ph1+ Ph1- What is Ph1 ? Cloning - the issues • The wheat genome is very large 17 Gb. (human 3Gb, yeast 0.12Gb) – Three closely related genomes! • No natural variation in Ph1 phenotype -Can’t create segregating populations, the starting point of all previous positional cloning projects • EMS treatments don’t yield mutants • But X-Ray and fast neutron irradiation do -A single deletion (ph1b) of the locus = 70Mb in size Defining the Ph1 locus Deletions Rice Deletions Griffiths et al 2006 Al-Kaff et al 2008 Deletions Wheat Brachypodium Defining the Ph1 locus further 2.5 Mb Al-Kaff et al. (2007) Cluster of 7 Cyclin dependent kinase-like (Cdks) genes on the long arm of 5B All defective genes = Ph1 locus Large segment of Heterochromatin inserted on polyploidisation Hypothesis- the defective 5B Cdk copies are suppressing the activity of the related Cdks elsewhere in the genome. But how to take the study further in wheat? Ph1 Cdk-like gene shows similarity to Cdk2 Cdk2 in mammals affects histone H1 phosphorylation So as a defective locus, does Ph1 suppress Cdk activity, hence histone H1 phosphorylation? Ph1 cdk+cyclinA compared to Cdk2+cyclinA Ph1-cdk gene Protein modeling Yousafzai and Al-kaff, 2010 Does Ph1 affect histone H1 phosphorylation? Human Histone H1 phosphorylation sites Cdk2 phosphorylates human histone H1 at consensus motifs (S/T) –P-X-K H11_HUMAN H1T_HUMAN H15_HUMAN H12_HUMAN H13_HUMAN H14_HUMAN Consensus (1) (1) (1) (1) (1) (1) (1) H11_HUMAN H1T_HUMAN H15_HUMAN H12_HUMAN H13_HUMAN H14_HUMAN Consensus (79) (80) (79) (76) (77) (76) (81) H11_HUMAN H1T_HUMAN H15_HUMAN H12_HUMAN H13_HUMAN H14_HUMAN Consensus (154) (151) (157) (156) (157) (156) (161) _ 1 80 MSETVPPAPAAS--AAPEKPLAGKKAKKPAKAAAASKKKPAGPSVSELIVQAASSSKERGGVSLAALKKALAAAGYDVEK MSETVPAASASAGVAAMEKLPTKKRGRKPAGLISAS-RKVPNLSVSKLITEALSVSQERVGMSLVALKKALAAAGYDVEK MSETAPAETATP--APVEKSPAKKKATKKAAGAGAAKRKATGPPVSELITKAVAASKERNGLSLAALKKALAAGGYDVEK MSETAPAAPAAA--PPAEKAPVKKKAAKKAGGTP---RKASGPPVSELITKAVAASKERSGVSLAALKKALAAAGYDVEK MSETAPLAPTIP--APAEKTPVKKKAKKAGATAGK--RKASGPPVSELITKAVAASKERSGVSLAALKKALAAAGYDVEK MSETAPAAPAAP--APAEKTPVKKKARKSAGAAK---RKASGPPVSELITKAVAASKERSGVSLAALKKALAAAGYDVEK MSETAPAAPAAP APAEKTPVKKKAKK AGAAGAS RKASGPPVSELITKAVAASKERSGVSLAALKKALAAAGYDVEK TPVK 81 160 NNSRIKLGIKSLVSKGTLVQTKGTGASGSFKLNKKASSVETKPGASKVATKT--KATGASKKLKKATGASK---KSVKTP NNSRIKLSLKSLVNKGILVQTRGTGASGSFKLSKKVIPKSTRSKAKKSVSAKTKKLVLSR-----DSKSPK----TAKTN NNSRIKLGLKSLVSKGTLVQTKGTGASGSFKLNKKAASGEAKPKAKKAGAAKAKKPAGAT--PKKAKKAAGAKKAVKKTP NNSRIKLGLKSLVSKGTLVQTKGTGASGSFKLNKKAASGEAKPKVKKAGGTKPKKPVGAAKKPKKAAGGATPKKSAKKTP NNSRIKLGLKSLVSKGTLVQTKGTGASGSFKLNKKAASGEGKPKAKKAGAAKPRKPAGAAKKPKKVAGAATPKKSIKKTP NNSRIKLGLKSLVSKGTLVQTKGTGASGSFKLNKKAASGEAKPKAKKAGAAKAKKPAGAAKKPKKATGAATPKKSAKKTP TPKK TPKK NNSRIKLGLKSLVSKGTLVQTKGTGASGSFKLNKKAASGEAKPKAKKAGAAK KKPAGAAKKPKKATGAATPKKSAKKTP 161 231 KKAKKPAATRKSSKNP---KKPKTVK-PKKVAKSPAKAKAVKPKAAKARVTKPKTAKPKKAAPKKK----KRAKKPRATTPKTVRS--GRKAKGAK-GKQQQKSPVKARASK-----SKLTQHHEVNVRKATSKK-----KKAKKPAAAGVKKVAK-SPKKAKAAAKPKKATKSPAKPKAVKPKAAKPKAAKPKAAKPKAAKAKKAAAKKK KKAKKPAAATVTKKVAKSPKKAKVAK-PKKAAKSAAKAVKP-------KAAKPKVVKPKKAAPKKK----KKVKKPATAAGTKKVAKSAKKVKTPQ-PKKAAKSPAKAKAPKPKAAKPKSGKPKVTKAKKAAPKKK----KKAKKPAAAAGAKKAK-SPKKAKAAK-PKKAPKSPAKAKAVKPKAAKPKTAKPKAAKPKKAAAKKK----KKAKKPAAAA TKK A SPKK SPKKAKAAK PKKAAKSPAKAKAVKPKAAKPKAAKPK AKPKKAAPKKK SPAK Is wheat histone H1 phosphorylated at Cdk2 consensus sites and is their phosphorylation altered by Ph1? Wheat histone H1 phosphorylated at Cdk2type consensus (S/T) –P-X-K sites Progenesis 0.08 0.06 0.04 0.02 PH W T 0.00 Cdk2-type phosphorylation on histone H1 is increased when Ph1 locus deleted Azahara Martinez, Ali Pendle, Alex Jones, Isabelle Colas Reduced homologous pairing, univalents homoeologous pairing Mutate or over-express Arabidopsis Cdkg Reduced homologous pairing, univalents 0 Ph1 copies Mutivalents Increased Cdk activity 2 Ph1 copies Bivalents Homologous pairing Homologous pairing 6 Ph1 copies Reduced Cdk activity Reduced homologous pairing, univalents John Doonan Greer et al. 2012 Reduced homologous pairing, univalents Moshe Feldman 1966 Metaphase I pairing • CDKG is closely related to Cdk2 and Ph1 • Mutant cdkg1 shows temperature-sensitive defects in synapsis and recombination of male meiosis Cdkg1 is partially asynaptic at 23oC Zheng et al. 2014 Asy1 Zyp1 DAPI Summary Deleting Ph1 increases Cdk activity- which increases histone H1 phosphorylation Result - pairing between homoeologous chromosomes Key question Can we mimic the effect of deleting Ph1 by increasing histone H1 phosphorylation and hence induce pairing between related chromosomes? Does increased Cdk-type activity induce pairing between related chromosomes? •Okadaic acid inhibits phosphatases •Okadaic acid increases histone H1 kinase activity •Does okadaic acid induce pairing between related chromosomes? Detached tiller method Okadaic acid induces pairing of related chromosomes in a wheat x rye hybrid No okadaic acid – mostly univalents Okadaic acid treatment produces a similar effect on chromosome pairing of related chromosomes as deleting Ph1 Knight et al., 2010 Okadaic acid - bivalents and other chromosome associations Wheat X Rye – Ph1 deleted Homoeologous pairing Does okadaic acid treatment affect the same Cdk2 consensus site as Ph1? YES! Progenesis 0.08 0.06 0.04 0.02 0.025 0.020 0.015 0.010 0.005 U O A _1 00 0.000 nt re at ed ratio phospho / non-phospho W PH T 0.00 The “Ph1” Cdk2-type consensus site shows increased phosphorylation with okadaic acid treatment Increased histone H1 phosphorylation leads to more “open” /decondensed chromatin? How does this affect pairing /recombination? Ph1 forms bivalents by eliminating multivalents Pachytene Zygotene Diplotene Diakinesis Leptotene Metaphase I Ph1+ 42 chromosomes Ph142 chromosomes Jenkins 1983 Holm, 1986,1988 high stringency synapsis but some multivalents at zygotene multivalents eliminated at pachytene 21 homologous bivalents at metaphase I multivalents lower stringency retained at synapsis with more pachytene multivalents at zygotene At both these stages condensation changes occur which would be affected by histone H1 phosphorylation What happens at the homologue recognition stage in wheat? homologues De-condensation /elongation of chromatin Rye segment Wheat telomeres homologous segments telomeres The identical chromosomes zip up from their telomere regions Pilar Prieto et al 2004 Nat Cell Biol In wheat- chromosomes remodel in both the presence and absence of Ph1 BUT there is asynchronous chromatin remodelling in the absence of Ph1 correlating with more incorrect associations at homologue recognition stage Ph1- Interstitial segments- 15% of the wheat chromosome Telomeres Ph1+ Ph1+ Pilar Prieto et al 2004 Nat Cell Biol Ph1+ Ph1+ De-condensation of chromosome segments is dependent upon their sequence similarity • Identical segments Segments elongated Synchronously before clustering 100% pairing • Similar segments Segments elongated but Not Synchronously 50% pairing • Distinct segments Reduced/Delayed 15% pairing Colas et al 2008 PNAS In wheat-rye hybrids without Ph1 homoeologous wheat-rye chromosomes only trigger a partial conformation change Diploidhomologues Hybrid- Ph1homoeologues Ph1- Hybrid- Ph1+ homoeologues Ph1+ Ph1- heterochromatin telomeres Pairing Some Pairing No Pairing Synapsis in diverged (related) chromosomes Homologues with divergent segments Chromosome segments remodel Little synaptic adjustment with Ph1 Telomeres Chromosome segments forming a circular structure Synaptic adjustment without Ph1 Recombination Colas et al., PNAS 2008 No recombination Synaptic Adjustment The Ph1 effect is important agronomically Wild species of wheat carry important traits for disease resistance and salt, cold and drought tolerance **Strategic Goal** Switch Ph1 on and off in elite wheat varieties crossed with wild species to introduce novel genes to the commercial crop Summary • Wheat is a global crop with a complex evolutionary history which gave it its hexaploid status • Ph1 stabilises the wheat genome by controlling pairing, and effectively turns it into a diploid • Ph1 is related to human Cdk2 which phosphorylates histone H1 and modifies chromatin conformation • Ph1 could be used to introduce novel genes into commercial crops Thanks Diolch Спасибо 37