* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download pathway_cell_models_2006
Cytokinesis wikipedia , lookup
Hedgehog signaling pathway wikipedia , lookup
Cellular differentiation wikipedia , lookup
Multi-state modeling of biomolecules wikipedia , lookup
Signal transduction wikipedia , lookup
Paracrine signalling wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
List of types of proteins wikipedia , lookup
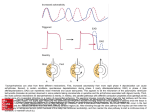
Biological systems and pathway analysis An introduction Protein-Protein Interactions How does the organisation of living matter in space and time give rise to biological processes? Post-genome informatics: Prediction of all interactions Synthesis gagccagcgagccag cgcgcgcgggcgggc ggacagatcggagcc gagcggggccgggcg gggcgctccctgcag + Systems Biology Emergence a behaviour or property resulting from interaction and self-organisation Biological systems are •modular (sub-systems) System •hierarchical •multiscale (space and time) Self-organisation Agent interactions Agents interacting components (modules), ongoing relationships, can evolve molecular complexes, pathways, networks, tissues transcription, catalysis, P-P interaction, cross-talk, cellcell signalling molecules pathways cells Global approaches: Systems Biology Perturbation Living cell Dynamic response time! Global approaches: Systems Biology Perturbation Living cell Dynamic response time! Biological organisation (e.g., gene families, structural genomics, pathways) “Virtual cell” -omics time series Global approaches: Systems Biology Perturbation Living cell Dynamic response time! Biological organisation (e.g., gene families, structural -omics time series genomics, pathways) Bioinformatics Mathematical modelling Simulation •Basic principles “Virtual cell” •Practical applications Dynamic Pathway Models • Forefront of the field of systems biology • Types – Inter/intra-cellular – Metabolic networks – Gene networks – Signal transduction networks • Two types of formalism appearing in the literature: – data mining e.g. genome expression at gene or protein level contribute to conceptualisations of pathways – simulations of established conceptualisations Dynamic models of cell signalling …from pathway interaction and molecular data Erk1/Erk2 Mapk Signaling pathway …to dynamic models of pathway function Schoeberl et al., 2002 Simulations: Dynamic Pathway Models Schoeberl et al., 2002, Nat. Biotech. 20: 370 • These have recently come to the forefront due to emergence of highthroughput technologies. • Composed of theorised/validated pathways with kinetic data attached to every connection - this enables one to simulate the change in concentrations of the components of the pathway over time given initial parameters. Response Models × Signalling Pathways Models? • Charasunti et al. (2004) – ODE model of the action of Gleevec on the Crk-1 pathway in Chronic Myeloid Leukaemia Dynamic biochemistry • Biomolecular interactions • Protein-ligand interactions • Metabolism and signal transduction • Databases and analysis tools • Metabolic simulation • Metabolic databases and simulation • Dynamic models of cell signalling Types of Modelling Methods • Stochastic approaches – Simple statistics – Bayesian Networks • Deterministic – Boolean networks • ODE approach – Iterations in a system • Classification/Clustering approaches – Support Vector Machines – Neural Networks • Hybrid Models – mixture of the above Ideker & Lauffenberger, 2003, TiB 21(6): 255-262 Pathway simulation and analysis software accessible from http://sbml.org/index.psp BASIS BioCharon Bio Sketch Pad BioSpreadsheet BioUML BSTLab CADLIVE CellDesigner Cellerator Cellware Cytoscape DBsolve Dizzy E-CELL ESS Gepasi Jarnac JDesigner JigCell JSIM JWS Karyote* libSBML MathSBML MOMA Monod NetBuilder PathArt PathScout ProcessDB* SBW SCIpath SigPath Simpathica StochSim STOCKS TeraSim Trelis Virtual Cell WinSCAMP www.ucl.ac.uk/oncology/MicroCore/ microcore.htm Microarrays, Protein Pathways and SCIpath Microarray Technology Measure mRNA levels as approximation of genome expression Pathway Information mRNA https://www.ucl.ac.uk/oncology/MicroCore/microcore.htm •Pathway builder SBML BioPAX •MicroExpress maps microarray and proteomic data to pathway nodes •External database links Dynamic information processing in the cancer cell ‘Pathway signatures’ Molecular signatures for pathways are needed (not only for single molecules) A p53 Pathway A protein in the pathway Upregulation of the protein’s gene e.g. this protein is upregulated in prostate cancer compared to normal cells Greater upregulation… Downregulation of the protein’s gene Greater downregulation… Leukaemia types Leukaemia types Biomedicine ‘after the human genome’ Patient Molecular basis of disease Current disease models Molecular building blocks genes proteins Biomedicine ‘after the human genome’ Patient Physiology Clinical data Molecular basis of disease Current disease models Molecular building blocks genes proteins Biomedicine ‘after the human genome’ Patient Complex disease models Disease manifestation in organs, tissues, cells Computational modelling Molecular building blocks genes proteins Molecular organisation Tumour Systems Modelling • Four major areas of focus: GROWTH RESPONSE ANGIOGENESIS PATHWAYS delivery flow – – – – Growth Treatment Response Pathway Angiogenesis • Levels of granularity and scope – Phenomenological mechanistic Growth Models • Stamatakos et al. (1998) Proliferating: – 3D simulation – Satisfactory agreement with in vitro experiments Mitosis: Necrotic Products: t = 240h G0 Phase: Necrosis or Apoptotic Irradiation starts at t = 168h Cellular Automata (CA) approaches Determination of local interaction rules G 1 S G2 M G0 N Cell death Stamatakos et al. 1998, 2001 Stamatakos et al… Flowchart for the response of a single tumour cell to irradiation. Stamatakos et al., 2001, IEEE Trans. Inf. Tech. Biomed. 5(4) Physiome project “Virtual human” Simulation of complex models of cells, tissues and organs •40 years of mathematical modeling of electrophysiology and tissue mechanics •New models will integrate large-scale gene expression profiles http://www.physiome.org/ Physiome project patient organ Anatomy and integrative function, electrical dynamics Vessels, circulatory flow, exchanges, energy metabolism cell Cell models, ion fluxes, action potential, molecules, functional genomics