* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download H/D exchanged - Docenti.unina
Survey
Document related concepts
Transcript
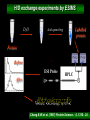
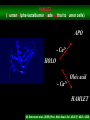
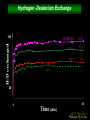
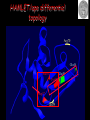
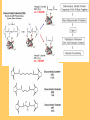
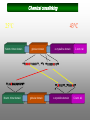
Protein structure depends on amino acid sequence and interactions Amino acid sequence Local interactions Long distance interactions Interactions between subunits Other methods? Tools for protein topology studies Complementary proteolysis Identification of the amide bond cleaved by a single proteolytic event that leads to complementary peptides Selective chemical modification Identification of the individual residue modified by, even aspecific mono-bifunctional, reagents Hydrogen/deuterium exchange Quantitative analysis of amide bonds accessible to solvent and available to exchange Sequence/Structure Paradigm higher order structures are the main determinants of the preferential cleavage site. peptide bonds buried in the protein core and/or located within rigid secondary structures are less accessible to proteases COMPLEMENTARY PROTEOLYSIS Proteolytic cleavage site HPLC ESMS ARG 93 -lactalbumin B Abs A Proteolysis under controlled conditions C C A A Exposed and flexible sites C B min Native protein RP-HPLC D Abs A Proteolysis under controlled conditions Conformational changes A D min Protein under different experimental conditions (ligands, denaturants, pH etc…) LC- MS LC- MS/MS Proteolysis Experiment Conditions • Proteases – Trypsin, Lys-C, Chymotrypsin, GluC, ………others • Buffer condition – pH, salt and detergent if necessary--Protein native condition? • Protein concentration • Time points – 5min, 10min, 30min, 1h, 2hrs and 4hrs. Complementary proteolysis H/D exchange and mass spectrometry Very fast R O R O N CH C N CH C H H Very slow Strongly depending on structural environment Factors that Slow Exchange Rate H/D-MS measures the rate at which peptide amide hydrogens exchange with the hydrogen in the water where the protein is dissolved. The rate of exchange reveals the degree of exposure of each amide hydrogen in the folded protein to water. In the unfolded portions, this rate is higher than that of structured, folded sections. Hydrogen bonding that creates the secondary structure, primarily alpha-helices and beta sheets. Protection from the solvent, primarily due to being buried in the hydrophobic core of the protein. Hydrogen bonding to water in the solvent (much smaller effect than the previous two). H/D exchange esperiments by ESIMS D2O Labelled protein Acid quenching Protein Before ESI Probe HPLC After Chung E.W. et al. (1997) Protein Science : 6, 1316 - 24 Labelled protein Pepsin digestion 5min, pH 2, 0°C ESI Probe HPLC Labelled peptides Before After Wang L. et al. (2002) Mol. Cell.Proteomics: 1, 132-138 HAMLET (Human Alpha-lactalbumin Made LEthal to Tumor cells) APO - Ca2+ HOLO - Ca2+ Oleic acid HAMLET M. Svensson et al. (2000) Proc. Natl. Acad. Sci. USA 97: 4221- 4226 Global H/D exchange profile HAMLET 0% 1759.77 100% 1777,27 apo 0% 1759.77 100% 1777,27 Intensity (%) 15 sec 1 min 5 min 15 min 60 min m/z m/z holo 0% 1759.77 100% 1777,27 Hydrogen -Deuterium Exchange 140 H/D exchanged HAMLET apo holo ~130 ~110 ~80 apo* 20 0 60 Time (min) Pepsin digestion for the local exchange profile A 1 310 KQFTKCELSQ LLKDIDGYGG 1 B IALPELICTM FHTSGYDTQA IVENNESTEY 3 4 2 310 61 CKSSQVPQSR NICDISCDKF 6 121 EKL 1 123 C LDDDITDDIM GLFQISNKLW CAKKILDIKG 60 5 310 D IDYWLAHKAL CTEKLEQWLC 3 120 Holo α-lactalbumin Apo α-actalbumin HAMLET 100 80 Holo α-lactalbumin is always less flexible than HAMLET and Apo 60 40 2 1 20 (12-23) (1-11+118-123) % exchanged protons 0 100 No substantial difference between HAMLET and Apo in peptides from the α domain 80 60 40 20 3 4 (24-31+103-107) (41-52) 0 100 80 60 6 40 (60-96) 5 20 (53-59) 0 0 20 40 60 00 time (min) 20 20 40 40 60 60 HAMLET incorporates a higher percentage of deuterium than Apo in peptides from the β domain HAMLET/apo differential topology Arg70 Glu46 Glu49 Phe53 Tyr50 Asp37 Gln39 Chemical crosslinking O O O O N O C CH2CH2CH2CH2CH2CH2C O O N O + NH3 + H3N Saccharomyces cerevisiae Hsp26 Hsp26 is an ATP independent cytosolic chaperone of Saccharomyces cerevisiae. Hsp26 expression is related to environmental stress (i.e. temperature). Hsp26 acts as a buffer for (partly) unfolded proteins, handing them over to other chaperon-systems, such as Hsp70. Hsp26 has the ability to form Dimers and large Oligomers of 24 subunits. Disassembling/Assembling of the oligomer plays a vital role in chaperon function ΔT substrate N I aggregate ΔT Hsp26 dissociated Hsp26 oligomer Hsp26-substrate complex N Chemical crosslinking EDAC 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide Lys O CHCl- NH2 CH3CH2 N C N Glu, Asp HO (CH2)3 N+ CH3 EDAC H N C O Legame isopeptidico CH3 in situ digestion MALDI-TOF analysis Identification of crosslinked peptides Chemical crosslinking 25°C 43°C N-term. trimer domain globular domain α-crystalline domain C-term. tail 116KDIDIEYHQNK126… 147VKVKESSSGK 156 158 151ESSSGKFK 23LLGEGGLRGYAPR35 190LKPQK194…198NHVKK20 2 N-term. trimer domain globular domain α-crystalline domain C-term. tail