* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Assignment: Amino Acids, Peptides, and Proteins
Magnesium transporter wikipedia , lookup
Butyric acid wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Citric acid cycle wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Western blot wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Metalloprotein wikipedia , lookup
Point mutation wikipedia , lookup
Peptide synthesis wikipedia , lookup
Proteolysis wikipedia , lookup
Genetic code wikipedia , lookup
Amino acid synthesis wikipedia , lookup
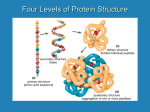
Assignment: Amino Acids, Peptides, and Proteins 1. What are the four amino acid classes? What structural criteria are used to assign the standard -amino acids into these classes? Provide examples. There are basically four different classes of amino acids determined by different side chains: (1) neutral non-polar, (2) neutral polar, (3) acidic, (4) basic. If amino acids side chain contains hydrocarbon alkyl groups (alkane branches) or aromatic (benzene rings), they are neutral non-polar acid amino. The term neutral is used because these R groups do not bear positive or negative charges because they interact poorly with water. Examples of aromatic form structure of nonpolar amino acids: glycine, valine, alanine, leucine, isoleucine Phenylalanine, Tryptophan, methionine, cystein, praline. If amino acids side chain contains a polar hydroxyl group, they are neutral polar amino acids. They are polar amino acids because they have functional groups capable of hydrogen bonding, and easily interact with water. The OH present enables them to participate in hydrogen bonding. Asparagine and glutamine are Amide (NH2) derivative of the acidic amino acids. Amide functional group of the acidic amino acids make the amino acid is highly polar. Examples of neutral polar amino acids: Serine, Threonine, Tyrosine, Asparagine, Glutamine If the amino acid side chain contains an acid functional group (carboxylate group), the whole amino acid produces an acidic solution, and they are acidic amino acids. Normally, an amino acid produces a nearly neutral solution since the acid group and the basic amine group on the root amino acid neutralize each other in the zwitterion. If the amino acid structure contains two acid groups and one amine group, there is a net acid producing effect. The two acidic amino acids are aspartic and glutamic (example). If the amino acids side chain contains an amine functional group, they are basic amino acid. The amino acid produces a basic solution because the extra amine group is not neutralized by the acid group. Amino acids which have basic side chains include: lysine, arginine, and histidine (examples). Amino acids with an amide on the side chain do not produce basic solutions i.e. asparagine and glutamine. 2. List and describe the diverse functions performed by proteins. Catalysis Structure Movement Defense Regulation Transport Storage Stress response 3. Describe the process of protein denaturation. What conditions cause denaturation? Denaturation is the loss of protein structure from quaternary to secondary but not to primary structure. Cause of denaturation Strong acids or base Organic solvents Detergents Reducing agents Salt concentrations Heavy metal ions Temperature changes Mechanical stress 4. Most amino acids appear bluish purple when treated with ninhydrin reagent. Proline and hydroxyproline appear yellow. Suggest a reason for the difference. Proline and hydroxyproline are both amino acids and do not lose their nitrogen atom when they react with ninhydrin. As a result, when proline reacts with ninhydrin, the following compound is formed This material is yellow. 5. Indicate the level(s) of protein structure to which each of the following contributes: a. amino acid sequence b. β-pleated sheet c. hydrogen bond d. disulfide bond a. The amino acid sequence is a polypeptide's primary structure. b. -pleated sheet is one type of secondary structure. c. Inter- and intrachain hydrogen bonds between N-H groups and carbonyl groups of peptide bonds are the principal feature of secondary structure. Hydrogen bonds formed between polar side chains are important in tertiary and quaternary structure. d. Disulfide bonds are strong covalent bonds that contribute to tertiary and quaternary structure. 6. What type of secondary structure would the following amino acid sequence be most likely to have? a. polyproline - left-handed helix b. polyglycine - β-pleated sheet c. Ala-Val-Ala-Val-Ala-Val - α-helix d. Gly-Ser-Gly-Ala-Gly-Ala - β-pleated sheet 7. List three factors that do not foster α-helix formation. The structural features of several amino acids do not foster -helix formation. Because the R group of glycine is too small the polypeptide chain becomes too flexible. Proline's rigid ring prevents the required rotation of the N-C bond. Sequences with larger numbers of amino acids with charged side chains (e.g., glutarnate) and bulky side chains (e.g., tryptophan) are also incompatible with helix formation. 8. Denaturation is the loss of protein function from structural change or chemical reaction. At what level of protein structure or through what chemical reaction does each of the following denaturation agents act? a. heat - hydrogen bonding, (secondary and tertiary structure) b. strong acid - hydrogen bonding (secondary and tertiary structure) and salt bridges (secondary and tertiary structure) c. saturated salt solution - salt bridges (tertiary structure) d. organic solvents (e.g., alcohol or chloroform) - hydrophobic interaction (tertiary structure) 9. A polypeptide has a high pI value. Suggest whichh amino acids might comprise it. Amino acids with basic side groups such as lysine, arginine, or tyrosine would contribute to a high pI value. 10. Outline the steps to isolate typical protein. What is achieved at each step? pg 152 First step: develop an assay for the protein of interest (must be specific) Must be convenient to perform because it will be used for many times Enzymes: disappearance of substrate or formation of product can be measured using spectrophotometer Nonenzymatic protein: detected by employing antibodies Extraction of proteins begins with cells disruption and differential centrifugation. Cells disruption: separated using homogenization method Differential centrifugation: using ultracentrifuge. Centrifuge to separate particles according to their size, surface area and relative density 11. Outline the steps to purify a protein. What criteria are used to evaluate purity? The first step in the isolation of a specific protein is the development of an assay which allows the investigator to detect it during the purification protocol. Next, the protein, as well as other substances, are released from source tissue by cell disruption and homogenization. Preliminary purification techniques include salting out, in which large amounts of salt are used to induce protein precipitation, and dialysis, in which salts and other low molecular weight material are removed. Further purification methods, which are adapted to each research effort at the discretion of the investigator, include various types of chromatography and electrophoresis. 12. List the types of chromatography used to purify proteins. Describe how each separation method works. (refer pg 152-153) Gel-filtration chromatography Gelatinous polymer separates molecules according to their size and shape they go through the pores Large molecules: excluded and move through the column quickly Smaller molecules: diffuse in and out of the pores, so their movement, through the column is retarded The smaller their molecular weight, the slower they move This different rates separate the protein mixture into bands, then collected separately Ion-exchange chromatography Separates proteins according to their charge Anion-exchange resins= bind reversibly with a protein negatively charged group Cation-exchange resins= bind to the positively charged The proteins that do not bind to the resin are removed Protein of interest, is recovered by an appropriate change in the solvent pH and/or salt concentration Affinity chromatography It uses a special noncovalent binding affinity between the protein and a special molecule (the ligand) After nonbinding protein molecules have passed through the column, the protein of interest is removed by altering the condition that affect binding 13. In sequencing a protein using carboxypeptidase, the protein is first broken down into smaller fragments, which are then separated from one another. Each fragment is then individually sequenced. If this initial fragmentation were not carried out, amino acid residues would build up in the reaction medium. How would these residues inhibit sequencing? In sequencing a polypeptide, the next residue in a sequence is determined by the increase in height of the next peak on the amino acid analyzer. If there is already a large amount of that amino acid present in the solution, it would be difficult, if not impossible, to detect any change in peak height. Small fragments have only a few amino acids, and this problem does not occur. 14. In an amino analysis, a large protein is broken down into overlapping fragments by using specific enzymes. Why must the sequences be overlapping? With overlapping fragments the segments can be fitted together because fragments that fit together have common sequences at their ends. if the segments are not overlapping, the order of the cannot be determined. 15. Hydrolysis of β-endorphin (a peptide containing 31 amino acid residues) produces the following amino acids: Tyr (1), Gly (3), Phe (2), Met, Thr (3), Ser (2), Lys (5), Gln (2), Pro, Leu (2), Val (2), Asn (2), Ala (2), Ile, His, and Glu Treatment with carboxypeptidase liberates Gln. Treatment with DNFB liberates DNP-Try. Treatment with trypsin produces the following peptides: Lys, Gly-Gln, Asn-Ala-Ile-Val-Lys, Try-Gly-Gly-Phe-Met- Thr-Ser-Glu-Lys, Asn-Ala-His-Lys, Ser-Gln-Thr-Pro-Leu-Val-Thr-Leu-Phe-Lys Treatment with chymotrypsin produces the following peptides: Lys-Asn-Ala-Ile-Val-Lys-Asn-AlaHis-Lys-Lys-Gly-Gln Tyr-Gly-Gly-Phe Met-Thr-Ser-Glu-Lys-Ser-Gln-Thr-ProLeu-Val-Thr-Leu-Phe What is the primary sequence of β-endorphin? The structure of -endorphin is Tyr-Gly-Gly-Phe-Met-Thr-Ser-Glu-Lys-Ser-Gln-Thr-Pro-LeuVal-Thr-Leu-Phe-Lys-Asn-Ala-Ile-Val-Lys-Asn-Ala-His-LysLys-Gly-Gln 16. Consider the following tripeptides: Gly-Ala-Val a. What is the approximate isoelectric point? The isoelectric point is calculated by taking the average of pKa values for the amino group of glycine (9.6) and the carboxyl group of valine (2.32) The answer is pl=5.96 b. In which direction will the tripeptide move when placed in an electric field at the following pH values? 1, 5, 10, 12 At pH 1, the tripeptide is positively charged and will move to the negative electrode. At pH 5 the tripeptide has a net zero charge and will not migrate. At pH 10 and 12, the tripeptide has a -1 charge and moves towards the positive electrode. 17. The following is the amino acid sequence of bradykinin, a peptide released by certain organisms in response to wasp stings: Arg-Pro-Pro-Gly-Phe-Ser-Pro-Phe-Arg What amino acids or peptides are produced when bradykinin is treated with each of the following reagents? a. carboxypeptidase b. chymotrypsin c. trypsin d. DNFB The following fragments are produced when bradykinin is treated with the indicated reagents: Carboxypeptidase – Arg and Arg-Pro-Pro-Gly-Phe-Ser-Pro-Phe Chymotrypsin – Arg-Pro-Pro-Gly-Phe, Ser-Pro-Phe, and Arg Trypsin – Arg and Pro-Pro-Gly-Phe-Ser-Pro-Phe-Arg DNFB - DNP-Arg and the following amino acid residues: 3 Pro, 1Gly, 2Phe, 1 Ser and 1 Arg