* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Quantitative inheritance
Gene expression profiling wikipedia , lookup
Public health genomics wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Genomic imprinting wikipedia , lookup
Gene expression programming wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Genome (book) wikipedia , lookup
Human genetic variation wikipedia , lookup
Genetic drift wikipedia , lookup
Selective breeding wikipedia , lookup
Designer baby wikipedia , lookup
Behavioural genetics wikipedia , lookup
Population genetics wikipedia , lookup
Microevolution wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Dominance (genetics) wikipedia , lookup
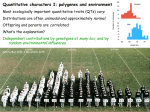
Quantitative inheritance Qualitative and Quantitative traits, Variation in traits, polygenic model Lecture 5 Aims • To equip students with knowledge on the inheritance of polygenic traits Learning Objectives • By the end of the topic, students should be able to • Differentiate quantitative traits from qualitative traits • Describe traits of importance in farm animals • Explain the polygenic model • Describe and understand the basis of quantitative variation and its measures • Describe terms values and means • Explain breeding values and genotypic values • Compute values for various terms DNA on chromosome (number per cell nucleus are species specific) Genes (control all traits in animals and plants) Qualitative (major) traits Quantitative (polygenic) traits Genotype (a collection of alleles an individual has in its genome) Interaction between alleles at a locus and between genes at different loci influence inheritance mode Phenotype (what the animal or plant looks like with respect to a particular trait or a composite combination of traits) Traits of interest in farm animals • Many traits in farm animals are measured on a continuous scale • And are termed quantitative traits • In contrast to traits that fall into distinct classes • Termed qualitative or discrete traits Types of traits Traits Qualitative Quantitative Controlled by single or few genes Each gene has a major effect Controlled by many genes, each with a small effect (influence) No environmental influence Influenced by environment Discrete in expression Continuous in expression Observed Measured rather than observed Described by proportions Described by statistics e.g. coat colour, hornness, shape of e.g. body weight, milk yield, ear (inherited in a simple mendelian egg production, fibre content, fashon) power Of economic importance Quantitative traits Are also called polygenic, controlled by many loci Each has little additive effect to the expression (animal performance) Are mostly complex in inheritance With many biochemical pathways and proteins, enzymes involved And measured on a continuous scale (as quantities) Polygenic model As the number of genes controlling a trait increases, the distribution of genetic effects become normal Quantitative traits are assumed to be controlled by many genes at many loci: thus the polygenic model This can be described by looking at the variation ~ shown in following slides Assumptions behind following slides • To illustrate discontinuous variation caused by genetic segregation as it is translated into continuous variation of metric traits 1. Consider any locus, each with two alleles ~ i.e. two forms of a gene, with equal gene frequency 2. There is no dominance of one allele at each locus 3. Then the dominant allele adds 1 unit to the measurement of a trait, recessive allele adds -1 unit 4. Loci are not linked, each loci having equal effect on phenotype (trait) measure In fact, all these assumptions are not true For a trait controlled by one locus genotypic value of quantitative trait~ single locus model Locus 1 with 2 alleles A, a Allele A has value +1; allele a has value -1 Genotypic classes and values are as below Genotypic class AA genotypic value 1 + 1 +2 Aa 1 + -1 0 aa -1 + -1 -2 Traits are discrete, comprised only few classes Example of determining genotypic value of quantitative trait, single locus Consider weaning weight in pigs at 15 weeks Mean weight = 20 kg Locus 1 with 2 alleles A, a Genotype Mean + genotypic value = weight AA 20 + 2 22 Aa 20 + 0 20 Aa 20 + 0 20 aa 20 + -2 18 With respect to locus 1 the pigs could be any of the three genotypes 2 2 1.5 1.5 Frequency Frequency Single locus model, only three classes 1 0.5 1 0.5 0 18 20 Value, wt 22 0 -2 0 Value 2 Two locus model Consider trait controlled by 2 Loci, each with 2 alleles A, a and B, b A and B have value +1; a and b values -1 Genotypic value for Bs, As as in above Genotype Genotype Genotypic value Weight AABB 24 AABb 22 AaBB 22 BB +2 Bb 0 AaBb 20 AAbb 20 bb -2 aaBB 20 Aabb 18 aabB 18 aabb 16 With combined effect of locus 1 & 2, the range of genotypes widens Two locus model AB Ab aB ab AB AABB AABb AaBB AaBb Ab AABb AAbb AaBb Aabb aB AaBB AaBb aaBB aaBb ab AaBb Aabb aaBb aabb Two locus model, genotype values AABB +4 AABb +2 AaBB +2 AaBb 0 AABb +2 AAbb 0 AaBb 0 Aabb -2 AaBB +2 AaBb 0 aaBB 0 aaBb -2 AaBb 0 Aabb -2 aaBb -2 aabb -4 Example of determining genotypic value of quantitative trait, two loci Consider Locus 2 with 2 alleles B, b AaBb AaBb aabB AaBb AaBB aabB AAbb AABb aabb Aabb aaBB 16 AABb 18 20 5 Frequency Aabb AaBb 6 3 2 AaBB AABB 22 4 24 1 0 16 18 20 22 24 Weight -4 -2 0 Genetic effects +2 +4 -4 -2 0 +2 Trait starts to appear quantitative +4 Example of determining genotypic value of quantitative trait, three loci Consider Locus 3 with 2 alleles C, c With combined effect of locus 1, 2 & 3, the range of genotypes and phenotypes widens even further Majority animals tend to be at the middle of the range and the distribution of the phenotype tend to be almost normal Number of genotype combinations n 2n Number of gene pairs with 2 alleles Number of possible gametes 3n Number of possible genotypes alleles A & a has n = 1; therefore, 2 possible gametes A & a; 3 possible genotypes AA, Aa & aa Check excel for genotypes and gametes Possible gametes and genotypes~ Can be illustrated through Punnet Square • 2 alleles, 1 loci • ~ 3 genotypes possible 2 loci, each with 2 alleles ~ 9 genotypes possible A a A AA Aa a Aa aa AB Ab aB ab AB AABB AABb AaBB AaBb Ab AABb AAbb AaBb Aabb aB AaBB AaBb aaBB aaBb ab AaBb Aabb aaBb aabb Sharp rise in genotypes and gametes as number of loci increase 1200000 4E+09 3.5E+09 1000000 3E+09 800000 2.5E+09 600000 2E+09 1.5E+09 400000 1E+09 200000 500000000 0 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 Number of gene pairs No of gametes No of genotypes Frequency Example of determining genotypic value of quantitative trait, three loci 20 18 16 14 12 10 8 6 4 2 0 14 16 18 20 22 24 26 Weight -6 -4 -2 0 +2 +4 +6 Work out 27 genotypes Quantitative traits are affected by more than three loci, thus produce a smooth normal distribution Assignment • Work out four loci model and determine • Genotype values • Frequency for each genotype • Plot the distribution Polygenic model As the trait get controlled by numerous loci, each with multiple alleles The number of genotypic classes becomes large, and the genotypic values become normally distributed How are these traits inherited The combined effect of the alleles at many loci make up genotypic value of an individual Genotypic value = the value of the animal’s genes to its own performance During reproduction, an animal passes an allele from each locus to its offspring (progeny) Which one is passed to is at random This makes each offspring to inherit a different set of alleles from the other Thus each offspring will have different genotypic value due to this random sampling of alleles from each parent – the Mendelian sampling How are these traits inherited It is this transmission of alleles from parents to offspring and the different set of combination that leads to observed variation in a trait Assumptions behind not always true • Consider any locus, each with two alleles ~ i.e. two forms of a gene, with equal gene frequency • Fact, > 2 alleles exist, each with different frequencies • Dominance effect takes place • • Loci can be linked Each loci can be with different effect • There is no dominance of one allele at each locus • Then the dominant allele adds 1 unit to the measurement of a trait, recessive allele adds -1 unit Loci are not linked, each loci having equal effect on phenotype (trait) measure • Check for effect of dominance in Animal Breeding software~ Inheritance of quantitative traits Continuous variation expressed • Variation of this sort, without natural discontinuities, is called Continuous variation • Characters or traits that exhibit continuous variation are called Quantitative traits or metric characters – Since their study depend on measurement instead of counting • The brach of genetics concerned with metric characters is called Quantitative or biometrical genetics • Since segregation of genes cannot be followed individually, mode of inheritance is called Quantitative inheritance Summary: polygenic model As the number of genes controlling a trait increases, the distribution of genetic effects become normal Quantitative traits are assumed to be controlled by many genes at many loci: thus the polygenic model Effect of the environment An individual deviates from its genotypic value due to environmental effects These include Nutrition Climate Disease Random measurement error Environmental effects The effect of the environment on a quantitative trait for a group of animals also tend to be normally distributed This assumes random environmental effects. Systematic environmental effects, e.g. year, season, housing differences, feed type, sex, etc can be accounted for Phenotypes An individual inherits genetic effects and receives environmental effects The combined effect of these make up the phenotype Phenotype is therefore, the sum of genetic effects and environmental effects Phenotype Genotype Environment P GE Phenotypic effects If P is expressed as deviation from group mean An individual’s deviation in phenotype is due to the sum of its deviation in genetic and environmental effects Phenotypic effects Animal Population Mean, kg G E P 1 40 +6 +6 52 2 40 +2 +14 56 3 40 -2 -4 34 4 40 +6 -2 44 5 40 -8 +4 36 Mean = average value of genes all individuals in a population have in common, plus the average level of management Phenotypic effects 8 4 2 0 -2 1 2 3 4 20 5 Phenotypic effects Genotypic value 6 -4 -6 -8 -10 Animal 15 10 5 0 1 2 3 Environmental deviations -5 16 14 -10 12 Animal 10 8 6 4 2 0 -2 -4 1 2 3 -6 Animal 4 5 4 5 All effects Phenotypic variation When a large number of animals are measured, you get variation in the phenotypic values for a trait Just as P = G + E, Variance in P is the sum of variance in genotypic effects and variance in environmental effects Phenotypic variation Due to large number of effects on the trait, that is why variation distribution is normal When you put all the effects together, you get normally distributed observations Measures of variation 100 80 70 60 Frequency As quantitative traits are (usually) normally distributed They can be described by the measure of the central tendency (mean) and its variance (standard deviation) 90 50 40 30 20 10 0 350 375 400 425 450 475 500 Weaning weight 525 550 575 600 625 More Normality: a review • It is bell curve • One of many distributions • Majority observations tend to be at the middle • Fewer observations are at extreme ends • Most traits in livestock are normally distributed Summary on variation in quantitative traits Quantitative traits show a normal distribution Due to Underlying genetic distribution, attributed to polygenic trait model An underlying environmental distribution It is the genetic variance in quantitative traits that is of importance in animal breeding It allows selection for genetically superior animals Properties of a population with metric characters • In quantitative traits, properties we can observe include • Means • Variances • Co-variances Based on these properties • Variance can be split into components • Through natural subdivision of population into families • And use that to measure degree of resemblance between relative • So that observations made on population can be used to predict outcome of a breeding method • And observe impact of a breeding program • And also to find out how observable properties of a population are influenced by genetic and nongenetic factors Concept of mean and variance Mean measures the central tendency of a population for a particular trait Variance and standard deviation describes the spread of a normally distributed population Mean X n X n Variance (Vx) Standard deviation (σ) Vx X X n 1 Vx 2 An example Live weight data for adult local sheep Check in excel An exercise Calculate mean and variance in excel For heart girth in sheep And for the data on pig production at our Students’ Farm ~ you will collect from the Animal Recording Technician Standard deviation (σ), what is it? σ=1 Describes the spread of the distribution σ=5 The greater the σ, the greater the spread ~68% of the measurements are ±1 σ units σ=10 Fewer measurements ~1% are greater than ±3 σ from the mean units Mean and variance Weaning Weight Frequency 100 80 60 40 20 Mean = ~525 More 625 600 575 550 525 500 475 450 425 400 375 350 0 standard deviation = ~50 •Mean •Describes the middle of a normal distribution •Standard deviation describes a width of a normal distribution Practically expect Variance to differ between flocks More variance in flocks kept in a wide environment like ranch than those kept under uniform environment like a barn More variance in local than improved strains More variance in unselected than in animals that have undergone selection V p VG VE V p VG VE V p VG VE VG VA VD VI Practically expect Practically expect Different flocks can have same mean but different variance Sakhula flock SF flock Mean, g 2.2 2.2 Variance, g2 0.04 0.36 Std, g 0.20 0.60 Simple properties of variance Adding a constant to each observation (measurement) does not change the variance, only the mean If X and Y are independent, Var X Y Var( X ) Var(Y ) E.g. Genetic and environmental effects, where there is no GxE interaction Two unrelated traits, where they are not genetically or environmentally correlated Further reading • Inheritance of quantitative traits • In Animal Breeding software Further explanation on terms • ‘Values’ and ‘means’ • ‘Average effects’ and ‘breeding values’ • Refer to Falconer Chapter 7 • And, Inheritance of quantitative traits • In Animal Breeding software • To be covered in detail in Applied Animal Breeding Summary of terms: Value • Value is basically what was expressed as Genotype value • Recall • Much as we measure the P, our interest is the G P GE V p VG VE Value • Recall genotypes and their frequencies under HW equilibrium Genotype Frequency Gij=Genotypic expression Genotypic value Yij=Phenotypic expression AA p2 G11 +a Y11 Aa 2pq G12 d Y12 aa q2 G22 -a Y22 Mean genotypic value of two homozygotes • Is equal to zero • Genotypic value is defined as deviation of the phenotype from the average of the two homozygous phenotypes • Recall, this is the value of genes to the animal itself – Mid-point between two homozygotes serves as an origin with zero value • Hence, genotypic values are measured as deviation from the mid-point A2A2 -a A1A2 0 d A1A1 Genotype +a Genotypic values Genotypic value of heterozygote ~ d • Determines the degree of dominance • When • d=0, means there is no dominance • Genes are just additive (additive case) A2A2 -a A1A2 0 d A1A1 Genotype +a Genotypic values Genotypic value of heterozygote ~ d • Determines the degree of dominance • When • d=a, means there is complete dominance • A1 is dominant over A2 ~ d is positive • A2 is dominant over A1 ~ d is negative A2A2 -a 0 A1A2 A2A2 -a -d 0 A1A2 A1A1 Genotype +a d Genotypic values A1A1 Genotype +a Genotypic values Genotypic value of heterozygote ~ d • Determines the degree of dominance • When • 0<d<a, means there is partial dominance A2A2 -a 0 A1A2 A1A1 Genotype d +a Genotypic values Genotypic value of heterozygote ~ d • Determines the degree of dominance • When • d>a, means there is over dominance A1A1 A1A2 A2A2 -a • The degree of dominance • Is expressed as +a 0 d d a d Genotype Genotypic values Example 7.1 in Falconer • Go through Example 7.1 in Falconer Mean Genotype AA Aa aa Frequency Gij=Genotypic expression p2 2pq q2 Genotypic value Yij=Phenotypic expression +a d -a Y11 Y12 Y22 G11 G12 G22 • Mean genotypic value is obtained by f G ij ij • Where • Fij is genotype frequency • Gij if genotypic value Mean Genotype Frequency Genotypic value Freq x Value AA Aa aa p2 2pq q2 +a d -a p2a 2pqd -q2a a(p-q)+2dpq f G ij ij mean sumofcolumns •Note that the sum is the mean since the sum is divided by 1, 1 being equal to sum of three genotype frequencies •i.e. p2+2pq+q2 = 1 Derivation of the mean fijGij p a 2 pqd q (a) 2 2 p a q a 2 pqd 2 2 a p q 2 pqd 2 2 a p q p q 2 pqd a p q 2 pqd Contribution of any locus to population mean • Includes – A term associated with homozygotes – a(p-q) – A term associated with heterozygotes – 2pqd • Check Falconer for contribution and mean under • • • • No dominance Complete dominance When A1 were fixed in a population (p=1) When A2 were fixed in a population (q=1) Contribution to Mean from many loci • Assuming no epistasis a p q 2 pqd Go through example 7.1 and 7.2 in Falconer • Note • Under normal environment • Mean environmental deviation is zero • Hence, population mean refers to phenotypic or genotypic values • Values are in unit of measure • Check excel to see how gene frequency influences the mean Go through example 7.1 in Falconer Example 7.1 in Falconer Weight, g Mean under differing gene frequencie s, p Deviation Mean 0.5 AA Actual measure Aa aa Mean a Values d (a) 14 12 6 10 4 2 -4 1 14 14 4 9 5 5 -5 2.5 14 14 14 4 15 3 4 4 4 9 9 9 5 5 5 -5 6 -6 -5 -5 -5 -2.5 3 -3 Complete, positive 11.5 dominance Complete, negative 6.5 dominance 12 Over dominance 6 Over dominance 0 No dominance, mean determined 10 by gene frequency 14 10 6 10 4 0 -4 11 Partial dominance Average effect and breeding value • With respect to transmission of value from parents to offspring • Parents pass their genes and not genotypes • Genotypes are created afresh in each generation • Genotype values are therefore, not transmitted • A new measure of values that refer to genes and not genotypes is needed – This new value associated with genes and not genotypes is the • Average effect Basis of inheritance Average effect of a particular gene (allele) • The mean from the population mean of individuals which received that allele from one parent, the allele received from the other parent having come at random from the population • This new value allows us to assign a breeding value to individuals Breeding value • A value associated with the genes carried by an individual and transmitted to its offspring • This is value of genes to the animal offspring • That is, genotypic value of animal’s future progeny Breeding value • Since it is only genes that are transmitted to offspring, • It is therefore, the average effects of parents’ genes that determine the mean genotypic value of its progeny • The value of the individual, judged by the mean value of its progeny is the breeding value of the individual Breeding value and Genetic value • Genetic value • Value of genes to the individual itself • Includes dominant deviation that is not passed to offspring • Since each passes one allele • Breeding value • Value of genes to progeny • Sum of average effects of the individual’s alleles carried • Average effect of an allele depends on frequency of the allele in a population Breeding value, example • Under a single locus model • If αA1= 10 and αA2= -10 • BVA1A1= 10 + 10 = • BVA1A2= 10 + -10 = • BVA2A2= -10 + -10 = 20 0 -20 • Breeding values = the sum of the average effect of the individual’s alleles (α) Breeding value, example • Under a single locus model If αA1= 5 and αA2= -2 • BVA1A1= 5 + 5 • BVA1A2= 5 + -2 • BVA2A2= -5 + -5 = = = 10 3 -10 • Breeding values = the sum of the average effect of the individual’s alleles (α) G, BV and dominance • Since BVs are sum of average effects of genes • The heterozygote is always halfway between the two homozygotes • Irrespective of dominance • with no dominance • Genetic and breeding values are equal • With some dominance • Genetic and breeding values differ • See details in Excel Breeding value terms • Also referred to as • Additive genotype • Variation in breeding values also referred to as • Additive effects of genes • Breeding value in livestock is the genetic merit of an animal, used to rank animals and is an aid to selection • The difference between genotypic value, G and breeding value, A of a particular genotype is known as dominance deviation, D Summary • Breeding values are expressed as a deviation of the population mean • with the population mean dependent on genotypic values and frequencies • dominance G With no dominance G=A, with dominance G≠A • Animals with a rare allele will have a larger breeding value • either positive or negative BV Summary • Breeding values are additive • BVs are used to predict progeny performance • By estimating expected genetic value coming from one parent used in breeding • Most likely the sires ~ details in software on Inheritance of animal breeding Point to note • Breeding values always halved when predicting progeny performance • Referred to as transmitting ability • Remember • BV represents the sum of average effects of two alleles – Only one of which is passed on • These concepts relating genetic to BV could be extended from a single locus model to a multiple locus model