* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lec. 26 - Genomics
Gene expression wikipedia , lookup
Whole genome sequencing wikipedia , lookup
Gene regulatory network wikipedia , lookup
Community fingerprinting wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Genetic engineering wikipedia , lookup
Genomic imprinting wikipedia , lookup
Ridge (biology) wikipedia , lookup
Genomic library wikipedia , lookup
Non-coding DNA wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Gene expression profiling wikipedia , lookup
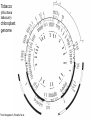
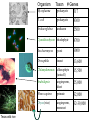
Genomes & The Tree of Life • Archaea - small circular genome • Prokarya - small to very small (e.g., Mycobacterium) circular genomes • Eukarya - 3 genomes – Mitochondrial – small to micro-sized, linear and circular, prokaryotic origin – Chloroplast – small, circular, prokaryotic origin – Nucleus – large, linear chromosomes; evidence of archaea, prokaryotic and “protoeukaryotic” origins Chloroplast DNA in Green Plants 1. Circular, multi-copy (20100/organelle) 2. ~160,000 bp; ~125 genes 3. Most genes of two types: • Photosynthesis • Genetic functions (mostly translation) Tobacco (Nicotiana tabacum) chloroplast genome From Kloppstech, Westhof et al. Plant nuclear genome sizes are large and widely varied. x 1000 to get bp Lilium longiflorum (Easter lily) = 90,000 Mb Fritillaria assyriaca (butterfly) = 124,900 Mb Protopterus aethiopicus (lungfish) = 139,000 Mb What about genetic complexity? How many genes do organisms have? Organism Texas wild rice Taxon Mycoplasma # Genes prokaryote 517 E. coli prokaryote 4300 Archaeoglobus archaeon 2500 Cyanidioschyzon rhodophyte 4700 Saccharomyces yeast 6000 Drosophila insect 13,600 Chlamydomonas chlorophyte (unicell) 15,500 Arabidopsis angiosperm, dicot 25,000 Homo sapiens primate 32,000 Oryza (rice) angiosperm, monocot 32-39,000 Mycoplasma : How many genes essential for growth (under lab conditions)? • Using transposon mutagenesis, ~150 of the 517 genes could be knocked out; ~ 300 genes deemed essential (under lab conditions), which included: – ~100 of unknown function – Genes for glycolysis & ATP synthesis – ABC transporters – Genes for DNA replication, transcription and translation Science 286, 2165 (1999) Genomic and species differences contributing to the wide range of nuclear genome sizes There can be great variation in the: 1. Fraction of highly repeated DNA 2. Abundance of "Selfish DNA“ (transposons, etc.) 3. Frequency and sizes of introns – Humans have many & larger introns 4. Genetic redundancy Genetic Redundancy • The sizes of many gene families has increased in some organisms more than others • Accounts at least partially for the relatively high genetic complexity of plants. Genetic Redundancy or Duplication yeast Drosophila Arabidopsis No. of genes 6200 13,600 25,000 No. of gene families 4380 8065 11,000 1820 5535 14,000 No. of genes from duplication Impact of Horizontal Transfer on Genomes • ~ 20% of the E. coli genome was obtained by lateral transfer. • Viral and bacterial pathogens can transfer DNA from host to host. • Some nuclear genes came from organellar genomes (some relatively recently). • Selfish DNAs such as mobile introns and transposons occasionally transfer horizontally. What can you do with whole genomes & sequences? 1. Predict much about the functions of a poorly studied or difficult organism - only ~1-5% of bacteria in the environment are culturable Transport and metabolic pathways of the Lyme disease spirochaete, Borrelia predicted from the genome sequence. Nature 390, 583 What can you do with whole genomes & sequences? 1. Predict much about the functions of a poorly studied or difficult organism. 2. Can examine genome-wide expression patterns with microarrays (e.g., cancer versus normal cells). Can immobilize 1,000-5,000 DNAs (genes) on one microarray glass slide. 1. Hybridize slide to cDNAs that were obtained by reverse transcription from total mRNAs with a fluorescent nucleotide. 2. Scan slide with a laser and process fluorescent image. Can simultaneously compare 2 different mRNA preparations by using different colored fluorescent nucleotides. Red- induced mRNA Green- decreased mRNA Yellow – unchanged mRNA What can you do with whole genome sequences? 1. 2. 3. 4. 5. Predict much about the functions of a poorly studied or difficult organism. Can examine genome-wide expression patterns with microarrays (e.g., cancer v. normal cells). Identify new drug targets. More rapidly identify genes linked to a trait. Rapidly identify a gene for an identified protein by mass spectrometry – compare mass spectrum of the protein with the predicted patterns from all of the genes of a sequenced genome (Proteomics).