* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Chapter 2: The Chemical Context of Life
Maximum parsimony (phylogenetics) wikipedia , lookup
Mitochondrial DNA wikipedia , lookup
Genetic engineering wikipedia , lookup
DNA barcoding wikipedia , lookup
Genomic imprinting wikipedia , lookup
Metagenomics wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Ridge (biology) wikipedia , lookup
Gene expression programming wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Gene expression profiling wikipedia , lookup
Pathogenomics wikipedia , lookup
Designer baby wikipedia , lookup
Transitional fossil wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Genome (book) wikipedia , lookup
History of genetic engineering wikipedia , lookup
Minimal genome wikipedia , lookup
Computational phylogenetics wikipedia , lookup
Genome evolution wikipedia , lookup
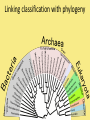
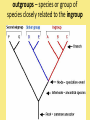
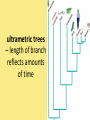
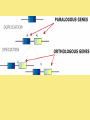
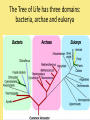
Chapter 25: Phylogeny and Systematics phylogeny – evolutionary history of a species or group of species systematics – analytical approach to understanding the diversity and relationships of present and past organisms Phylogenies based on common ancestors using fossil, morphological and molecular evidence the fossil record – based mostly on the sequence in which fossils have accumulate in sedimentary rock strata (layers) Sedimentary rock forms when silt builds up on bottom of waterway. More deposited on top, compress older sediments into rock. morphological and molecular homologies • organisms with similar morphology or similar DNA closely related analogy is not homology; analogy is a similarity due to convergent evolution (organisms adapting to similar environment/niche but no common ancestor) • distinguishing analogy from homology critical to constructing phylogenies homoplasies – analogous structures that have evolved independently Marsupial mole (Australia) No recent common ancestor eutherian mole (North America) molecular homologies • compare nucleic acids of two species; if very similar, organisms closely related • a hard to tell how long ago they shared a common ancestor; must also look at fossil record • mathematical tools can distinguish between distant homologies from coincidental matches Systematics connects classification with evolutionary history taxonomy – ordered division of organisms into categories bionomial nomenclature – scientific name • binomial – 2- part scientific name developed by Linnaeus “Linnean system” • genus – first part of name • specific epithet – second part of name Homo sapiens = humans, means “wise man” Heirarchical classification • • • • • • • • Domain Kingdom Phylum Class Order Family Genus Species mnemonic to help you remember: “Dreadful King Phillip Came Over From Great Spain” Linking classification with phylogeny phylogenetic trees – branching diagrams that depict hypotheses about evolutionary relationships. • uses groups nested within more inclusive groups • constructed from series of dichotomies (2-way branch points) • each branch point represents a divergence of two species from a common ancestor cladogram – shows patterns of shared characteristics clade – group of species that includes an ancestral species and all of its descendants cladistics – analysis of how species grouped into clades • clades can be nested inside larger clades – ex. cat family within a larger clade that includes dog family monophyletic group – ancestral species and all of its descendants paraphyletic group– when we lack information about some members of the clade polyphyletic group– several species that lack a common ancestor (need more work to uncover species that tie them together into a monophyletic clade) Shared Characteristics – types of homologous similarities Chordate characteristics Shared primitive character – shared beyond the taxon. Shared derived character – evolutionary novelty unique to that clade. Ex. hair only found in mammals Why morphology alone does not show evolutionary relationship: • Closely related organisms not always similar in appearance (rapid environmental change leads to rapid evolution; also, small changes in genes can lead to large morphological differences) • Organisms that appear similar not always closely related (convergent evolution) • Just because 2 groups share primitive characters does not mean they are closely related outgroups – species or group of species closely related to the ingroup • less closely related than members of the ingroup • have a shared primitive character that predates both ingroup and outgroup members phylogenetic trees – show estimated time since divergence • chronology of a phylogenetic tree is relative; not absolute phylograms – length of a branch reflects number of changes in a DNA sequence ultrametric trees – length of branch reflects amounts of time maximum parsimony “Occam’s Razor” – first investigate the simplest explanation that is consistent with the facts • Aim is to find the shortest tree that has the smallest number of changes The top tree has the most parsimony maximum likelihood – given certain rules of how DNA changes over time, a tree can be found that reflects the most likely sequence of evolutionary events Often the most parsimonious tree is also the most likely phylogenic trees are hypotheses of how the organisms are related to each other • Best hypothesis is one that fits all the available data • May be modified when new evidence introduced • Sometimes there is compelling evidence that the best hypothesis is not the most parsimonious Organisms’ genomes document their evolutionary history • importance of studying rRNA and mitochondrial DNA (mtDNA) rRNA changes very slowly; used to study divergences that happened a very long time ago mtDNA changes very rapidly; used to study divergences that happened recently • – useful for studying relationships between groups of humans ex. how Native Americans descend from Asian population that crossed the Bering Land Bridge 13,000 years ago Gene duplication one of most important types of mutation in evolution because it increases # of genes in genome. • can lead to further evolutionary changes gene families – groups of related genes in an organism’s genome • result of repeated duplications • have a common ancestor orthologous genes – homologous genes passed in a straight line from one generation to the next. • can diverge only after speciation • can be found in separate gene pools due to speciation paralogous genes – result of gene duplication. Found in more than one copy of the same genome • can diverge in the same gene pool Genome evolution • Orthologous genes are widespread and can extend over huge evolutionary distances • 99% of genes in humans and mice are orthologous; 50% of genes in humans and yeast are orthologous – demonstrates that all living organisms share many biochemical and developmental pathways Molecular clocks – way of measuring absolute time of evolutionary change • based on some genes seem to evolve at a constant rate • # of nucleotide substitutions in orthologous genes is proportional to time elapsed since the species branched from common ancestor neutral theory – for genes that change regularly enough to use as a molecular clock, these changes are probably a result of genetic drift and are mostly neutral (neither adaptive nor detrimental) • conclusion: much evolutionary change has no effect on fitness; therefore, not influenced by selection. • most new mutations are harmful and therefore removed quickly The universal tree of life • Genetic code universal to all forms of life so all life must share a common ancestor • Researchers trying to link all organisms in a “tree of life” • use rRNA genes for this; as they evolve most slowly The Tree of Life has three domains: bacteria, archae and eukarya The early history of these domains is not yet clear • due to horizontal gene transfer, substantial interchanges of genes between organisms of different domains • horizontal gene transfer due to transposable elements and fusion of different organisms (first eukaryote fusion of ancient bacteria with ancient archae)