* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Globular proteins
Biochemistry wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein moonlighting wikipedia , lookup
Western blot wikipedia , lookup
Protein domain wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Implicit solvation wikipedia , lookup
Protein folding wikipedia , lookup
Metalloprotein wikipedia , lookup
Homology modeling wikipedia , lookup
Proteolysis wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein adsorption wikipedia , lookup
Protein structure prediction wikipedia , lookup
X-ray crystallography wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
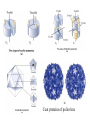
The first tertiary structure known was that of Myoglobin by John Kendrew (1950) It provided several clues about the globular protein structure. It is a small (Mr 16,700) oxygen binding protein in muscle cells 70% amino acid residues are in a helical form, The peptide backbone is made up of relatively straight a helices joined by bends and b turns. Three of the 4 proline residues are present at the bends. The clues: 1. The nonpolar residues Val, Leu, Ile, Met, and Phe largly occur in the interior of the protein away from the aqueous solvent layer 2. The charged polar residues Arg, Lys, His, Asp, and Glu are largely located at the surface in contact with aqueous solvent 3. The uncharged polar groups Ser, Thr, Asn, Gln, Tyr and Trp are usually on protein surface and are also found inside of the protein where they are always hydrogen bonded. Forces which stabilize the protein structure: Ionic interactions: Little contribution towards stability of native structure Dipole-Dipole interaction: Week forces but contribute to stability Hydrogen Bonding Forces: Very important role in 3o structure Hydrophobic Forces: Very Important contribution Disulfide bonds: Very important Techniques used for the determination of three dimensional structure 1. Circular Dichroism: 2. X-ray crystallography 3. NMR spectroscopy Circular Dicroism (CD): When polymers of optically active monomers are exposed to circularly polarized light, the absorptivty is different for right and left-circularly polarized light. The difference in the molar absorptivity varies with the wave length of the light. The CD spectra obtained for various secondary structures gives a characteristic pattern. X-Ray Crystallography: When a beam of X-ray of a given wave length falls on a crystal, the xrays are diffracted by the electrons of various atoms of the crystal. The diffracted X-rays are recorded on a photographic film or x-ray film by producing a pattern of spots with various intensities. By analysis of the x-ray diffraction pattern, spacing between different atoms of the molecule can be determined. X-ray diffraction pattern can give an idea about the electron densities in three dimensional space. Many proteins have been crystallized and their structures have been determined by x-ray diffraction pattern. Originally the process of calculations for atomic lattices from the diffraction pattern was very laborious but now through the development of computer software it has become more feasible. NMR Spectroscopy: Nuclear magnetic resonance is a manifestation of nuclear spin angular momentum of atomic nuclei. Only certain atoms including 1h, 13C, 15N, 19F and 31P posses the spin which can give rise to NMR signals. Nuclear spin generates a magnetic dipole which can either be in the direction of an applied static magnetic field (parellel, low energy level) or in anti-parallel direction (high energy level). When a suitable frequency of electromagnetic energy is applied at the right angle of the nuclei aligned in the magnetic field, it can absorb some energy and go to high energy level. The absorption spectrum thus obtained reflect the nature of the nuclei and its immediate electronic environment. With the advent of two dimensional NMR, distance dependent coupling of nuclear spins in nearby atoms through space (nuclear Overhause effect, NOSY) or the coupling of the nuclear spins of the atoms connected by covalent bonds (total correlation spectroscop, TOSY)can be measured. Supersecondary structures: Also called motifs or simply folds Particularly stable arrangement of several elements of secondary structure and connection between them Some simple motifs: 1. b-a-b loop and a-a corner can create layers in which the inner sides are shielded from water. 2. b-hairpin motif 3. b-barrels: Twisted b sheet conformation are more stable and they roll up to form b-barrels. The b barrel (a single domain of a hemolysin From bacterium S.aureus Quaternary Structure: Many proteins have multiple polypeptide subunits. It may be heteromeric (with different types of mono-meric subunits) or a multimer of same kind of subunits. The repeating structural units in a multimeric protein is called protomer. Identical subunits of a multimer proteins are generally arranged in a few ways of symmetric patterns. Cyclic symmetry: involving rotation around a single axis Dihedral symmetry: Two fold rotational axix intersects an n-fold (4fold in picture) axis at right angle. Other complex rotational symmetries are possible e.g. icosahedral symmetry The quaternary structure of deoxyhemoglobin Coat proteins of poliovirus