* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Protein-Chemistry_Svar-lektionsuppgifter
Protein mass spectrometry wikipedia , lookup
Immunoprecipitation wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Protein design wikipedia , lookup
Protein structure prediction wikipedia , lookup
Protein–protein interaction wikipedia , lookup
List of types of proteins wikipedia , lookup
Bimolecular fluorescence complementation wikipedia , lookup
Protein folding wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein purification wikipedia , lookup
Western blot wikipedia , lookup
Circular dichroism wikipedia , lookup
Metalloprotein wikipedia , lookup
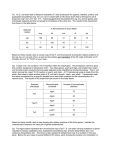
Answers to lesson exercises 1 a) kcat/knooncat = 4.6 x 103/0.5x10-6 s-1/s-1M-1 = 9.2x109 M = Ceff (Effective concentration) b) Enzyme catalyzed reactions are monomolecular, while this uncatalyzed reaction is bimolecular. The reacting groups are close to one another with correct orientation in the active site, which is entropically favorable. c) kcat/KM = 4.6x103/1,2x10-4 = 3.8x107 M-1s-1< kdiff = 108-109 M-1s-1 The rate is almost as high as the diffusion limited rate (kcat/KM is the rate constant by which E and S combine to form products at low concentrations of S.) d) 2) a) The reason to excite at 295 nm is to obtain a specific contribution from the tryptophans. At 280 nm also Tyr and Phe contribute to the fluorescence. b) The Trp mutant that gives the most blue-shifted spectrum is the Trp that is the most exposed. The reason to this is that the removed Trp must have been red-shifted, i.e. exposed and absence of one exposed Trp will give rise to a blue-shifted spectrum. Thus, Trp94 is the most exposed Trp in barnase. c) The reason to the increased fluorescence intensity when Trp 94 is mutated must depend on energy-transfer to this Trp, which in turn is quenched. d) The emission is increased when His18 mutated to a Gly despite the fact that none of these are fluourescent amino acids. The reason must be that His18 is a quencher of one of the three Trp:s, likely Trp94 (see c)) 3) See the Scatchard plot below: Scatchard formula: r/[A] = nKa – Ka r, where r = molar bound ligand/ [protein]total And [protein]total = 1.2/60 000 = 2.0x10-5 M [A] = free ligand concentration, i.e. the concentration of cAMP on the outside of the dialysis tube. Molar bound ligand is calculated as the difference in concentration between cAMP on the inside and the outside of the dialysis tube. Outside (M) Inside (M) Bound ligand (M) r r/[A] -6 -6 -6 0.7x10 2.7x10 2.0x10 0.1 1.4x105 -6 -6 -6 2.3x10 8.3x10 6.0x10 0.3 1.3 x105 4.5x10-6 1.5x10-5 1.1x10-5 0.5 1.2 x105 -5 -5 -5 1.0x10 2.6x10 1.6x10 0.8 0.8 x105 -5 -5 -5 3.0x10 5.4x10 2,4x10 1.2 0.4 x105 7.0x10-5 9.8x10-5 2.8x10-5 1.4 0.8 x105 -4 -4 -5 1.5x10 1.8x10 3.0x10 1.5 0.1 x105 Kd = 1/Ka Ka = the slope of the curve = 1x105 Thus, Kd = 1x10-5 M n = 1.6 The reason to a non-integer number of binding sites may be a) that the molecular weight determined by ultracentrifugation may not be quite exact b) a bad preparation of protein with a certain amount of denatured protein which does not bind cAMP. 4) E TyrAMP E TyrAMP# E TyrATP E Tyr C35G ΔΔG (kcal/mol) β (=) 1.65 1.24 1.24/1.65 = 0.75 0 0 0 0 H48G ΔΔG (kcal/mol) β (=) 1.83 1.61 1.61/1.83=0.9 0.81 0.81/1.83=0.4 0.37 0.37/1.83=0.2 Solid line: the wild-type Dashed line: the C35G mutant C35 does not affect the binding of the substrate, but affects the transistion state almost as much (75 %) as the final product. Thus, C35 is a catalytic group. H48 has a binding function to a certain degree, but above all H48 has a catalytic function.