* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Ab initio modelling tutorial (part II)
Implicit solvation wikipedia , lookup
X-ray crystallography wikipedia , lookup
Protein domain wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Protein purification wikipedia , lookup
Western blot wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Homology modeling wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
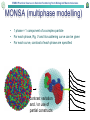
EMBO Practical Course on Solution Scattering from Biological Macromolecules Ab initio tutorial (part II) • Ambimeter – model independent ambiguity score • Damclust – alternative to Damaver • Gasbor (local & online) – real/reciprocal space • Monsa (online) – Nucleoprotein – Dimeric complex • Dammin – limited search volume – anisometry restraint AMBIMETER: tool for Ambiguity Assessment Map of renormalized scattering profiles from exhaustive set of shape topologies is employed to quantitatively evaluate the ambiguity of a small-angle-scattering curve Map of SAXS profiles density Extreme cases 0.0 Flat Disc Rod Sphere lg(I/I0) 0 10 100 1000 10000 -0.4 -0.8 -1.2 -1.6 1 2 3 4 AMBIMETER is now a part of SASFLOW pipeline 5 s*Rg 6 18 Skeletons with identical SAXS curves DamClust: Assessment of multimodality (has damaver & friends inside) Creates the complete graph by iteratively• joining the clusters (singles) Selects the optimal threshold as a compromise between the number of clusters and averaged spread within the cluster Clustering of multiple SAS models – Discrepancies (distances) between multiple models as criteria for grouping – Normalized spatial deviation serves as a distance between heterogeneous models (e.g. bead models) – R.m.s.d. is employed for those with atom-to-atom correspondence (e.g. rigid body models) EMBO Practical Course on Solution Scattering from Biological Macromolecules Dummy Residues Model • Proteins typically consist of folded polypeptide chains composed of amino acid residues • At a resolution of 0.5 nm each amino acid can be represented as one entity (dummy residue) • In GASBOR a protein is represented by an ensemble of K DRs those are – Identical – Have no ordinal number – For simplicity are centered at the Cα positions EMBO Practical Course on Solution Scattering from Biological Macromolecules n: 0 1 2 3 4 5 6 Fib(n): 1 1 2 3 5 8 13 21 34 55 89 144 233 377 610 987 7 ∆ρ=0.03 e/A3 Width=3A ρ=0.334 e/A3 8 9 10 11 12 13 14 15 EMBO Practical Course on Solution Scattering from Biological Macromolecules MONSA (multiphase modelling) • 1 phase = 1 component of a complex particle • For each phase, Rg, V and its scattering curve can be given • For each curve, contrast of each phase are specified contrast variation and / or use of partial constructs EMBO Practical Course on Solution Scattering from Biological Macromolecules Monsa case1: protein-RNA complex 252 AA protein (two domains) 67 nucleotides RNA Three curves in total: Free RNA Complex Free protein EMBO Practical Course on Solution Scattering from Biological Macromolecules Monsa case2: dimeric compex • 172 AA (20kDa) and 538 AA (70 kDa) proteins • Dimerization via small one • Two-fold symmetry axis EMBO Practical Course on Solution Scattering from Biological Macromolecules Dammin features missing in Dammif • Specific (limited) search volume (sphere, ellipsoid, cylinder, parallelepiped, User supplied) • Icosahedral and cubic symmetries • Anisometry restraint EMBO Practical Course on Solution Scattering from Biological Macromolecules Anisometry (direction) • both shapes are symmetric and extended