* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download yeast - chem.uwec.edu
Protein design wikipedia , lookup
Protein folding wikipedia , lookup
List of types of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Homology modeling wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein purification wikipedia , lookup
Protein domain wikipedia , lookup
Western blot wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
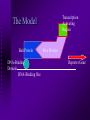
The Yeast Two-Hybrid System Anne C. Luebke What is the yeast two-hybrid system used for? Identifies novel protein-protein interactions Can identify protein cascades Identifies mutations that affect proteinprotein binding Can identify interfering proteins in known interactions (Reverse Two-Hybrid System) How does it work? Uses yeast as a model for eukaryotic protein interactions A library is screened or a protein is characterized using a bait construct Interactions are identified by the transcription of reporter genes Positives are selected using differential media Transcription Activating Region The Model Bait Protein DNA-Binding Domain DNA-Binding Site Prey Protein Reporter Gene Steps to Screen a Library Create the Bait Plasmid Construct from the gene of interest and the DNA binding domain of Gal4 or LexA or other suitable domain Transform with the bait construct a yeast strain lacking the promoter for the reporter genes and select for transformed yeast Transform the yeast again with the library plasmids Select for interaction Sequence analysis Isolate plasmid from yeast and transform E. coli Purify plasmid from E. coli and sequence Blast sequence against database for known proteins or construct a possible protein sequence from the DNA sequence and compare to other proteins Reporter Genes LacZ reporter - Blue/White Screening HIS3 reporter - Screen on His+ media (usually need to add 3AT to increase selectivity) LEU2 reporter - Screen on Leu+ media ADE2 reporter - Screen on Ade+ media URA3 reporter - Screen on Ura+ media (can do negative selection by adding FOA) Plasmid Constructs Plasmids are constructed with the Gal4 DNA binding domain (or other suitable domain) in front of a Multiple Cloning Site (MCS) The plasmid contains genes that can be used for selection such as Amp, Leu2, Ura3, or Trp1 Sample Plasmid From Golemis Lab Homepage False Positives False positives are the largest problem with the yeast two-hybrid system Can be caused by: Non-specific binding of the prey Ability to induce transcription without interaction with the bait (Majority of false positives) Elimination of False Positives Sequence Analysis Plasmid Loss Assays Retransformation of both strain with bait plasmid and strain without bait plasmid Test for interaction with an unrelated protein as bait Two (or more) step selections Advantages Immediate availability of the cloned gene of the interacting protein Only a single plasmid construction is required Interactions are detected in vivo Weak, transient interactions can be detected Can accumulate a weak signal over time Examples of Uses of the Yeast Two-Hybrid System Identification of caspase substrates Interaction of Calmodulin and L-Isoaspartyl Methyltransferase Genetic characterization of mutations in E2F1 Peptide hormone-receptor interactions Pha-4 interactions in C. elegans References Bartel, Paul, C. Chien, R. Sternglanz, S. Fields. “Elimination of False Positives that Arise in Using the Two-Hybrid System.” Biotechniques (1993) Vol. 14, no. 6, p. 920-924. Chien, Cheng-ting, P. Bartel, R. Sternglanz, S. Fields. “The two-hybrid system: A method to identify and clone genes for proteins that interact with a protein of interest.” Proc. Natl. Acad. Sci. USA (1991) Vol. 88, p. 9578-9582. Fields, Stanley, O. Song. "A novel genetic system to detect protein-protein interactions." Nature (1989) Vol. 340, p.245-246. James, Philip, J. Halladay, E. Craig. "Genomic Libraries and a Host Strain Designed for Highly Efficient Two-Hybrid Selection in Yeast." Genetics (1996) Vol. 144, p. 1425-1436. Kamada, S, H. Kusano, H. Fujita, M. Ohtsu, R. Koya, N. Kuzumaki, Y. Tsujimoto. "A cloning method for caspase substrates that uses the yeast two-hybrid system: Cloning of the antiapoptotic gene gelsolin." Proc. Natl. Acad. Sci. USA (1998) Vol 95, p. 8532-8537. O'Connor, Mirriam, C. O'Connor. "Complex Interactions of the Protein L-Isoaspartyl Methyltransferase and Calmodulin Revealed with the Yeast Two-hybrid System." The Journal of Biological Chemistry (1998) Vol. 273, p. 12909-12913. Staudinger, Jeff, J. Zhou, R. Burgess, S. Elledge, E. Olson. "PICK1: A Perinuclear Binding Protein and Substrate for Protein Kinase C Isolated by the Yeast Two-Hybrid System." The Journal of Cell Biology (1995) Vol. 128, p. 263-271. References continued Vidal, Marc, P. Braun, E. Chen, J. Boeke, E. Harlow. "Genetic Characterization of a mammalian protein-protein interaction domain by using a yeast reverse two-hybrid system." Proc. Natl. Acad. Sci. USA (1996) Vol. 93, p. 1032110326. White, Michael. "The yeast two-hybrid system: Forward and reverse." Proc. Natl. Acad. Sci. USA (1996) Vol 93, p. 10001-10003. Zhu, Jianwei, C. Kahn. "Analysis of a peptide hormone-receptor interaction in the yeast two-hybrid system." Proc. Natl. Acad. Sci. USA (1997) Vol. 94, p. 13063-13068. Lab of Erica Golemis http://www.fccc.edu/research/labs/golemis/EG_homepage.html Special thanks to Dr. Susan Mango and the University of Utah