* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download CHAPTER 4 Proteins: Structure, Function, Folding
Survey
Document related concepts
Transcript
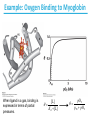
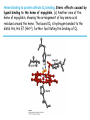
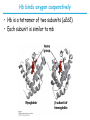
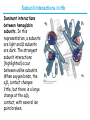
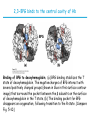
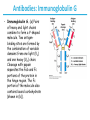
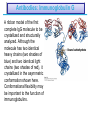
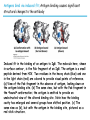
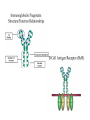
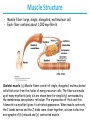
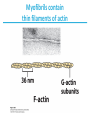
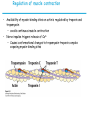
5| Function of Globular Proteins; and now the exciting stuff! CHAPTER 5: Function of Globular Proteins Key topics in protein function: • Reversible binding of ligands is essential – Specificity of ligands and binding sites – Ligand binding is often coupled to conformational changes, sometimes quite dramatic (Induced Fit) – In multisubunit proteins, conformational changes in one subunit can affect the others (Cooperativity) – Interactions can be regulated • Illustrated examples by: – Hemoglobin (Hb) antibodies (Ab), and muscle contraction Interaction with Other Molecules • Reversible, transient process of chemical equilibrium: A + B AB • A molecule that binds to a protein is called a ligand – Typically a small molecule • A region in the protein where the ligand binds is called the binding site • Ligand binds via same noncovalent forces that dictate protein structure (see Chapter 4) – Allows the interactions to be transient Binding: Quantitative Description • Consider a process in which a ligand (L) binds reversibly to a site in a protein (P) P • + ka L PL kd The kinetics of such a process is described by: – the association rate constant ka or the dissociation rate constant kd • After some time, the process will reach the equilibrium where the association and dissociation rates are equal • The equilibrium composition is characterized by the equilibrium constant Ka ka [P] [L] kd [PL] ka [PL] Ka [ P ] [ L] k d Binding: Analysis in Terms of the Bound Fraction • In practice, we can often determine the fraction of occupied binding sites (θ) • Substituting [PL] with Ka[L][P], we’ll eliminate [PL] [PL] [PL] [P] K a [L][P] K a [L][P] [P] • Eliminating [P] and rearranging gives the result in terms of equilibrium assoc. constant • In terms of the more commonly used equilibrium dissoc. constant [ L] [ L] K d [ L] [ L] 1 Ka Binding: Graphical Analysis • The fraction of bound sites depends on the free ligand concentration and Kd • Experimentally – Ligand concentration is known – Kd can be determined graphically or via – least-squares regression Graphical representations of ligand binding. The fraction of ligand-binding sites occupied, θ, is plotted against the concentration of free ligand. Both curves are rectangular hyperbolas. (a) A hypothetical binding curve for a ligand L. The [L] at which half of the available ligand-binding sites are occupied is equivalent to 1/Ka, or Kd. The curve has a horizontal asymptote at θ = 1 and a vertical asymptote (not shown) at [L] = –1/Ka. [ L] [ L] K d [L] [L]total Example: Oxygen Binding to Myoglobin When ligand is a gas, binding is expressed in terms of partial pressures. [L] K d [L] pO 2 p50 pO 2 For previous slide: • Myoglobin. The eight α-helical segments (shown here as cylinders) are labeled A through H. Nonhelical residues in the bends that connect them are labeled AB, CD, EF, and so forth, indicating the segments they interconnect. A few bends, including BC and DE, are abrupt and do not contain any residues; these are not normally labeled. (The short segment visible between D and E is an artifact of the computer representation.) The heme is bound in a pocket made up largely of the E and F helices, although amino acid residues from other segments of the protein also participate. • Graphical representations of ligand binding. The fraction of ligand-binding sites occupied, θ, is plotted against the concentration of free ligand. Both curves are rectangular hyperbolas. (b) A curve describing the binding of oxygen to myoglobin. The partial pressure of O2 in the air above the solution is expressed in kilopascals (kPa). Oxygen binds tightly to myoglobin, with a P50 of only 0.26 kPa. Binding: Thermodynamic Connections • Interaction strength can be expressed as – association (binding) constant Ka, units M-1 – dissociation constant Kd, units M, Kd = 1/Ka – interaction (binding) free energy Go, units: kJ/mol Definitions – Go = Ho -TSo : enthalpy and entropy – Ka = [PL]/[P][L] Kd = [P][L]/[PL] • Relationships – Go = -RT ln Ka = RT ln Kd (RT at 25oC is 2.48 kJ/mol) • Magnitudes – Strong binding: Kd < 10 nM – Weak binding: Kd > 10 M Examples of Binding Strength Specificity: Lock-and-Key Model • Proteins typically have high specificity: only certain ligands bind • High specificity can be explained by the complementary of the binding site and the ligand. • Complementary in – size, – shape, – charge, – or hydrophobic/hydrophilic character • “Lock and Key” model by Emil Fisher (1894) assumes that complementary surfaces are preformed. + Specificity: Induced Fit • Conformational changes may occur upon ligand binding (Daniel Koshland in 1958) – This adaptation is called the induced fit – Induced fit allows for tighter binding of the ligand – Induced fit allows for high affinity for different ligands • Both the ligand and the protein can change their conformations + Globins are oxygen-binding proteins • Protein side chains lack affinity for O2 • Some transition metals bind O2 well but would generate free radicals if free in solution • Organometallic compounds such as heme are more suitable, but Fe2+ in free heme could be oxidized to Fe3+ • Solution – Capture the oxygen molecule with heme that is protein bound – Myoglobin is the main oxygen storage protein – Hemoglobin is a circulating oxygen-binding protein Structures of Porphyrin and Heme • Heme. The heme group is present in myoglobin, hemoglobin, and many other proteins, designated heme proteins. Heme consists of a complex organic ring structure, protoporphyrin IX, with a bound iron atom in its ferrous (Fe2+) state. (a) Porphyrins, of which protoporphyrin IX is only one example, consist of four pyrrole rings linked by methene bridges, with substitutions at one or more of the positions denoted X. (b, c) Two representations of heme (derived from PDB ID 1CCR). The iron atom of heme has six coordination bonds: four in the plane of, and bonded to, the flat porphyrin ring system, and (d) two perpendicular to it. Structure of Myoglobin. The eight α-helical segments (shown here as cylinders) are labeled A through H. Nonhelical residues in the bends that connect them are labeled AB, CD, EF, and so forth, indicating the segments they interconnect. A few bends, including BC and DE, are abrupt and do not contain any residues; these are not normally labeled. (The short segment visible between D and E is an artifact of the computer representation.) The heme is bound in a pocket made up largely of the E and F helices, although amino acid residues fromother segments of the protein also participate. The heme group viewed from the side. This view shows the two coordination bonds to Fe2+ that are perpendicular to the porphyrin ring system. One is occupied by a His residue, sometimes called the proximal His; the other is the binding site for oxygen. The remaining four coordination bonds are in the plane of, and bonded to, the flat porphyrin ring system. Binding of Carbon Monoxide • CO has similar size and shape to O2; it can fit to the same binding site • CO binds over 20,000 times better than O2 because the carbon in CO has a filled lone electron pair that can be donated to vacant d-orbitals on the Fe2+ • Protein pocket decreases affinity for CO, but it still binds about 250 times better than oxygen • CO is highly toxic as it competes with oxygen. It blocks the function of myoglobin, hemoglobin, and mitochondrial cytochromes that are involved in oxidative phosphorylation CO vs. O2 Binding to Free Heme Steric effects caused by ligand binding to the heme of myoglobin. (a) Oxygen binds to heme with the O2 axis at an angle, a binding conformation readily accommodated by myoglobin. (b) Carbon monoxide binds to free heme with the CO axis perpendicular to the plane of the porphyrin ring. When binding to the heme in myoglobin, CO is forced to adopt a slight angle because the perpendicular arrangement is sterically blocked by His E7, the distal His. This effect weakens the binding of CO to myoglobin. Heme binding to protein affects O2 binding. Steric effects caused by ligand binding to the heme of myoglobin. (c) Another view of the heme of myoglobin, showing the arrangement of key amino acid residues around the heme. The bound O2 is hydrogen-bonded to the distal His, His E7 (His64), further facilitating the binding of O2. Could myoglobin transport O2? pO2 in lungs is about 13 kPa: it sure binds oxygen well pO2 in tissues is about 4 kPa: it will not release it! Would lowering the affinity (P50) of myoglobin to oxygen help? Graphical representations of ligand binding. The fraction of ligand- binding sites occupied, θ, is plotted against the concentration of free ligand. Both curves are rectangular hyperbolas. (b) A curve describing the binding of oxygen to myoglobin. The partial pressure of O2 in the air above the solution is expressed in kilopascals (kPa). Oxygen binds tightly to myoglobin, with a P50 of only 0.26 kPa. For effective transport affinity must vary with • A sigmoid (cooperative) binding curve. A sigmoid binding curve can be viewed as a hybrid curve reflecting a transition from a low-affinity to a high-affinity state. Because of its cooperative binding, as manifested by a sigmoid binding curve, hemoglobin is more sensitive to the small differences in O2 concentration between the tissues and the lungs, allowing it to bind oxygen in the lungs (where pO2 is high) and release it in the tissues (where pO2 is low). pO2 How can affinity to oxygen change? • Must be a protein with multiple binding sites • Binding sites must be able to interact with each other • This phenomenon is called cooperativity – positive cooperativity • first binding event increases affinity at remaining sites • recognized by sigmoidal binding curves – negative cooperativity • first binding event reduces affinity at remaining sites The Hill Plot of Cooperativity • Hill plots for oxygen binding to myoglobin and hemoglobin. When nH 5 1, there is no evident cooperativity. The maximum degree of cooperativity observed for hemoglobin corresponds approximately to nH 5 3. Note that while this indicates a high level of cooperativity, nH is less than n, the number of O2-binding sites in hemoglobin. This is normal for a protein that exhibits allosteric binding behavior. Two Models of Cooperativity: Concerted vs.Sequential Two general models for the interconversion of inactive and active forms of a protein during cooperative ligand binding. Although the models may be applied to any protein— including any enzyme—that exhibits cooperative binding, we show here four subunits because the model was originally proposed for hemoglobin. (a) In the concerted, or all-or-none, model (MWC model), all subunits are postulated to be in the same conformation, either all (low affinity or inactive) or all (high affinity or active). Depending on the equilibrium, Keq, between and forms, the binding of one or more ligand molecules (L) will pull the equilibrium toward the form. Subunits with bound L are shaded. (b) In the sequential model, each individual subunit can be in either the or form. A very large number of conformations is thus possible. Cooperativity is a special case of allosteric regulation • Allosteric protein – Binding of a ligand to one site affects the binding properties of a different site, on the same protein – Can be positive or negative – Homotropic • Normal ligand of the protein is the allosteric regulator – Heterotropic • Different ligand affects binding of the normal ligand • Cooperativity = positive homotropic regulation Hb binds oxygen cooperatively • Hb is a tetramer of two subunits (a2b2) • Each subunit is similar to mb Sequence Similarity between Hemoglobin and Myoglobin For previous slide; The amino acid sequences of whale myoglobin and the α and β chains of human hemoglobin. Dashed lines mark helix boundaries. To align the sequences optimally, short gaps must be introduced into both Hb sequences where a few amino acids are present in the other, compared sequences. With the exception of the missing D helix in Hbα, this alignment permits the use of the helix lettering convention that emphasizes the common positioning of amino acid residues that are identical in all three structures (shaded). Residues shaded in pink are conserved in all known globins. Note that the common helix-letterand- number designation for amino acids does not necessarily correspond to a common position in the linear sequence of amino acids in the polypeptides. For example, the distal His residue is His E7 in all three structures, but corresponds to His64, His58, and His63 in the linear sequences of Mb, Hbα, and Hbβ, respectively. Nonhelical residues at the amino and carboxyl termini, beyond the first (A) and last (H) α-helical segments, are labeled NA and HC, respectively. Where we left off TUESDAY Subunit Interactions in Hb Dominant interactions between hemoglobin subunits. In this representation, α subunits are light and β subunits are dark. The strongest subunit interactions (highlighted) occur between unlike subunits. When oxygen binds, the α1β1 contact changes little, but there is a large change at the α1β2 contact, with several ion pairs broken. Subunit Interactions: Details; Some ion pairs that stabilize the T state of deoxyhemoglobin. (a) Close-up view of a portion of a deoxyhemoglobin molecule in the T state. Interactions between the ion pairs His HC3 and Asp FG1 of the β subunit (blue) and between Lys C5 of the α subunit (gray) and His HC3 (its α-carboxyl group) of the β subunit are shown with dashed lines. (Recall that HC3 is the carboxyl-terminal residue of the β subunit.) Subunit Interactions: Details; Some ion pairs that stabilize the T state of deoxyhemoglobin. (b) Interactions between these ion pairs, and between others not shown in (a), are schematized in this representation of the extended polypeptide chains of hemoglobin. R and T States of Hb • T = Tense state, – More interactions, more stable – Lower affinity for O2 • R = Relaxed state, – Fewer Interactions, more flexible – Higher affinity for O2 • O2 binding triggers a T R conformational change • Conformational change from the T state to the R state involves breaking ion pairs between the α1-2 interface R and T States of Hemoglobin The TR transition. In these depictions of deoxyhemoglobin, as in Figure 5–9, the β subunits are blue and the α subunits are gray. Positively charged side chains and chain termini involved in ion pairs are shown in blue, their negatively charged partners in red. The Lys C5 of each α subunit and Asp FG1 of each β subunit are visible but not labeled. Note that the molecule is oriented slightly differently than in Figure 5–9. The transition from the T state to the R state shifts the subunit pairs substantially, affecting certain ion pairs. Most noticeably, the His HC3 residues at the carboxyl termini of the β subunits, which are involved in ion pairs in the T state, rotate in the R state toward the center of the molecule, where they are no longer in ion pairs. Another dramatic result of the TR transition is a narrowing of the pocket between the β subunits. Conformational change is triggered by oxygen binding Changes in conformation near heme on O2 binding to deoxyhemoglobin. The shift in the position of helix F when heme binds O2 is thought to be one of the adjustments that triggers the T → R transition. pH Effect on O2 Binding to Hb • Actively metabolizing tissues generate H+, lowering the pH of the blood near the tissues relative to the lungs • Hb Affinity for oxygen depends on the pH – H+ binds to Hb and stabilizes the T state • Protonates His146 which then forms a salt bridge with Asp94 • Leads to the release of O2 (in the tissues) • The pH difference between lungs and metabolic tissues increases efficiency of the O2 transport • This is known as the Bohr effect pH Effect on O2 Binding to Hb • Effect of pH on oxygen binding to hemoglobin. The pH of blood is 7.6 in the lungs and 7.2 in the tissues. Experimental measurements on hemoglobin binding are often performed at pH 7.4. Hb and CO2 Export • CO2 is produced by metabolism in tissues and must be exported • 15–20% of CO2 is exported in the form of a carbamate on the amino terminal residues of each of the polypeptide subunits. • Notice: – the formation of a carbamate yields a proton which can contribute to the Bohr Effect – the carbamate forms additional salt bridges stabilizing the T state • The rest of the CO2 is exported as dissolved bicarbonate – Formed by carbonic anhydrase, and also producing a proton 2,3-Bisphosphoglycerate regulates O2 binding • Negative heterotropic regulator of Hb function • Present at mM concentrations in erythrocytes – Produced from an intermediate in glycolysis • Small negatively charged molecule, binds to the positively charged central cavity of Hb • Stabilizes the T states 2,3-BPG binds to the central cavity of Hb Binding of BPG to deoxyhemoglobin. (a) BPG binding stabilizes the T state of deoxyhemoglobin. The negativecharges of BPG interact with several positively charged groups (shown in blue in this surface contour image) that surround the pocket between the β subunits on the surface of deoxyhemoglobin in the T state. (b) The binding pocket for BPG disappears on oxygenation, following transition to the R state. (Compare Fig. 5–10.) 2,3-BPG allows for O2 release in the tissues and adaptation to changes in altitude • Effect of BPG on oxygen binding to hemoglobin. The BPG concentration in normal human blood is about 5 mM at sea level and about 8 mM at high altitudes. Note that hemoglobin binds to oxygen quite tightly when BPG is entirely absent, and the binding curve seems to be hyperbolic. In reality, the measured Hill coefficient for O2-binding cooperativity decreases only slightly (from 3 to about 2.5) when BPG is removed from hemoglobin, but the rising part of the sigmoid curve is confined to a very small region close to the origin. At sea level, hemoglobin is nearly saturated with O2 in the lungs, but just over 60% saturated in the tissues, so the amount of O2 released in the tissues is about 38% of the maximum that can be carried in the blood. At high altitudes, O2 delivery declines by about one-fourth, to 30% of maximum. An increase in BPG concentration, however, decreases the affinity of hemoglobin for O2, so approximately 37% of what can be carried is again delivered to the tissues. Spectroscopic Detection of Oxygen Binding to Myoglobin • The heme group is a strong chromophore that absorbs both in ultraviolet and visible range • Ferrous form (Fe2+ ) without oxygen has an intense Soret band at 429 nm • Oxygen binding alters the electronic properties of the heme, and shifts the position of the Soret band to 414 nm • Binding of oxygen can be monitored by UV-Vis spectrophotometry • Deoxyhemoglobin (in venous blood) appears purplish in color and oxyhemoglobin (in arterial blood) is red Sickle-cell anemia is due to a mutation in hemoglobin • Glu6 Val in the chain of Hb • The new Valine side chain can bind to a different Hb molecule to form a strand • This sickles the red blood cells • Untreated homozygous individuals generally die in childhood • Heterozygous individuals exhibit a resistance to malaria Formation of Hb Strands in Sickle-Cell Anemia Normal and sickle-cell hemoglobin. (a) Subtle differences between the conformations of hemoglobin A and hemoglobin S result from a single amino acid change in the β chains. (b) As a result of this change, deoxyhemoglobin S has a hydrophobic patch on its surface, which causes the molecules to aggregate into strands that align into insoluble fibers. Two Types of Immune Systems • Cellular immune system - targets own cells that have been infected - also clears up virus particles and infecting bacteria - key players: Macrophages, killer T cells (Tc), and inflammatory T cells (TH1) • Humoral “fluid” immune system - targets extracellular pathogens - can also recognize foreign proteins - makes soluble antibodies - keeps “memory” of past infections - key players: B-lymphocytes and helper T-cells (TH2) Cellular Immune System • Antibodies bind to fragments displayed on the surface of invading cells • Phagocytes: specialized cells that eat invaders • Macrophages: large phagocytes that ingest bacteria that are tagged by antibodies Humoral Immune System • Vertebrates also fight infections with soluble antibodies that specifically bind antigens – Antigens are substances that stimulate production of antibodies • • • • Typically macromolecular in nature Recognized as foreign by the immune system Coat proteins of bacteria and viruses Surface carbohydrates of cells or viruses – Antibodies are proteins that are produced by B cells and specifically bind to antigens • Binding will mark the antigen for destruction or interfere with its function • A given antibody will bind to a small region (epitope) of the antigen • One antigen can have several epitopes Antibodies: Immunoglobulin G • Composed of two heavy chains and two light chains • Composed of constant domains and variable domains • Light chains: one constant and one variable domain • Heavy chains: three constant and one variable domain • Variable domains of each chain make up antigenbinding site (two/antibody) • Variable domains contain regions that are hypervariable (specifically the antigen-binding site) • Confers high antigen specificity Antibodies: Immunoglobulin G • Immunoglobulin G. (a) Pairs of heavy and light chains combine to form a Y-shaped molecule. Two antigenbinding sites are formed by the combination of variable domains from one light (VL) and one heavy (VH) chain. Cleavage with papain separates the Fab and Fc portions of the protein in the hinge region. The Fc portion of the molecule also contains bound carbohydrate (shown in (b)). Antibodies: Immunoglobulin G A ribbon model of the first complete IgG molecule to be crystallized and structurally analyzed. Although the molecule has two identical heavy chains (two shades of blue) and two identical light chains (two shades of red), it crystallized in the asymmetric conformation shown here. Conformational flexibility may be important to the function of immunoglobulins. Antigens bind via induced fit; Antigen binding causes significant structural changes to the antibody Induced fit in the binding of an antigen to IgG. The molecule here, shown in surface contour, is the Fab fragment of an IgG. The antigen is a small peptide derived from HIV. Two residues in the heavy chain (blue) and one in the light chain (red) are colored to provide visual points of reference. (a) View of the Fab fragment in the absence of antigen, looking down on the antigen-binding site. (b) The same view, but with the Fab fragment in the “bound” conformation; the antigen is omitted to provide an unobstructed view of the altered binding site. Note how the binding cavity has enlarged and several groups have shifted position. (c) The same view as (b), but with the antigen in the binding site, pictured as a red stick structure. The BCR The membrane-bound form of an antibody may be called a surface immunoglobulin (sIg) or a membrane immunoglobulin (mIg). It is part of the B cell receptor (BCR), which allows a B cell to detect when a specific antigen is present in the body and triggers B cell activation. The BCR is composed of surface-bound IgD or IgM antibodies and associated Ig-α and Ig-β heterodimers, which are capable of signal transduction. A typical human B cell will have 50,000 to 100,000 antibodies bound to its surface. Upon antigen binding, they cluster in large patches, which can exceed 1 micrometer in diameter, on lipid rafts that isolate the BCRs from most other cell signaling receptors. These patches may improve the efficiency of the cellular immune response. In humans, the cell surface is bare around the B cell receptors for several hundred nanometers, which further isolates the BCRs from competing influences. Antibody specificity is an important analytical reagent Protein Interactions Modulated by Chemical Energy • Use of chemical energy (ATP) can cause conformational changes in proteins, generally required for their function • Especially in motor proteins – Control movement of cells and organelles within cells • Allows for spatial and temporal regulation of interactions Muscle Structure • Muscle fiber: large, single, elongated, multinuclear cell • Each fiber contains about 1,000 myofibrils Skeletal muscle. (a) Muscle fibers consist of single, elongated, multinucleated cells that arise from the fusion of many precursor cells. The fibers are made up of many myofibrils (only six are shown here for simplicity) surrounded by the membranous sarcoplasmic reticulum. The organization of thick and thin filaments in a myofibril gives it a striated appearance. When muscle contracts, the I bands narrow and the Z disks come closer together, as seen in electron micrographs of (b) relaxed and (c) contracted muscle. Myofibrils contain thick filaments of myosin Myofibrils contain thin filaments of actin Myosin thick filaments slide along actin thin filaments Myosin thick filaments slide along actin thin filaments From previous slide 5-29b,c Skeletal muscle. (a) Muscle fibers consist of single, elongated, multinucleated cells that arise from the fusion of many precursor cells. The fibers are made up of many myofibrils (only six are shown here for simplicity) surrounded by the membranous sarcoplasmic reticulum. The organization of thick and thin filaments in a myofibril gives it a striated appearance. When muscle contracts, the I bands narrow and the Z disks come closer together, as seen in electron micrographs of (b) relaxed and (c) contracted muscle. FIGURE 5–30a Muscle contraction. Thick filaments are bipolar structures created by the association of many myosin molecules. (a) Muscle contraction occurs by the sliding of the thick and thin filaments past each other so that the Z disks in neighboring I bands draw closer together. Actomyosin Cycle • Muscle contraction occurs through a series of conformational changes to protein structure due to binding, hydrolysis, and release of ATP and ADP • Cycle has four steps 1. ATP binds to myosin myosin dissociates from actin 2. ATP is hydrolyzed a conformational change of myosin 3. Myosin re-connects to the actin filament at a different location release of Pi 4. Release of Pi Power stroke where myosin returns to initial state; shifting actin filament relative to the myosin tail release of ADP Actomyosin Cycle • Molecular mechanism of muscle contraction. Conformational changes in the myosin head that are coupled to stages in the ATP hydrolytic cycle cause myosin to successively dissociate from one actin subunit, then associate with another farther along the actin filament. In this way the myosin heads slide along the thin filaments, drawing the thick filament array into the thin filament array (see Figure 530). Regulation of muscle contraction • • Availability of myosin-binding sites on actin is regulated by troponin and tropomyosin – avoids continuous muscle contraction Nerve impulse triggers release of Ca2+ – Causes conformational changes to tropomyosin-troponin complex exposing myosin-binding sites From pervious slide Regulation of muscle contraction by tropomyosin and troponin. Tropomyosin and troponin are bound to F-actin in the thin filaments. In the relaxed muscle, these two proteins are arranged around the actin filaments so as to block the binding sites for myosin. Tropomyosin is a two-stranded coiled coil of α helices, the same structural motif as in α-keratin (see Fig. 4–11 ). It forms head-to-tail polymers twisting around the two actin chains. Troponin is attached to the actin-tropomyosin complex at regular intervals of 38.5 nm. Troponin consists of three different subunits: I, C, and T. Troponin I prevents binding of the myosin head to actin; troponin C has a binding site for Ca2+; and troponin T links the entire troponin complex to tropomyosin. When the muscle receives a neural signal to initiate contraction, Ca2+ is released from the sarcoplasmic reticulum (see Fig. 5–29a) and binds to troponin C. This causes a conformational change in troponin C, which alters the positions of troponin I and tropomyosin so as to relieve the inhibition by troponin I and allow muscle contraction. In the words of Porky Pig, That’s all folks!