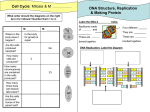
* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Exogenous nucleotides accelerate early replication
Non-coding DNA wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Cell culture wikipedia , lookup
DNA supercoil wikipedia , lookup
DNA vaccination wikipedia , lookup
Molecular cloning wikipedia , lookup
List of types of proteins wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Point mutation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Transformation (genetics) wikipedia , lookup
Vectors in gene therapy wikipedia , lookup