* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lecture 4| Enzyme Catalysis: Structural basis and energetics of
Western blot wikipedia , lookup
Epitranscriptome wikipedia , lookup
Vesicular monoamine transporter wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Drug design wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Restriction enzyme wikipedia , lookup
Biochemistry wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Ultrasensitivity wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Metalloprotein wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Ligand binding assay wikipedia , lookup
Nicotinamide adenine dinucleotide wikipedia , lookup
Biosynthesis wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Enzyme inhibitor wikipedia , lookup
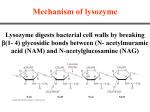
PHRM 836 September 8, 2015 Enzyme Catalysis: structural basis and energetics of catalysis Devlin, section 10.3 to 10.5 1. Enzyme binding of substrates and other ligands (binding sites, structural mobility) 2. Energe(cs along reac(on coordinate 3. Cofactors 4. Effect of pH on enzyme catalysis Enzyme catalysis: Review Devlin sections 10.6 and 10.7 • Defini(ons of catalysis, transi(on state, ac(va(on energy • Michaelis-‐Menten equa(on – Kine(c parameters in enzyme kine(cs (kcat, kcat/KM, Vmax, etc) – Lineweaver-‐Burk plot • Transi(on-‐state stabiliza(on • Meaning of proximity, orienta(on, strain, and electrosta(c stabiliza(on in enzyme catalysis • General acid/base catalysis • Covalent catalysis 2015, September 8 PHRM 836 -‐ Devlin Ch 10 2 Structure determines enzymatic catalysis as illustrated by this mechanism for ____ 2015, September 8 PHRM 836 -‐ Devlin Ch 10 www.studyblue.com 3 Substrate binding by enzymes • Highly complementary interac(ons between substrate and enzyme – Hydrophobic to hydrophobic – Hydrogen bonding – Favorable Coulombic interac(ons • Substrate binding typically involves some degree of conforma(onal change in the enzyme – Enzymes need to be flexible for substrate binding and catalysis. – Provides op(mal recogni(on of substrates – Brings cataly(cally important residues to the right posi(on. 2015, September 8 PHRM 836 -‐ Devlin Ch 10 4 Substrate binding by enzymes • Highly complementary interac(ons between substrate and enzyme – Hydrophobic to hydrophobic – Hydrogen bonding – Favorable Coulombic interac(ons • Substrate binding typically involves some degree of conforma(onal change in the enzyme – Enzymes need to be flexible for substrate binding and catalysis. – Provides op(mal recogni(on of substrates – Brings cataly(cally important residues to the right posi(on. Induced fit: promoted by rotation around bonds 2015, September 8 PHRM 836 -‐ Devlin Ch 10 Figure 10.11 Orotate phosphoribosyltransferase 5 Substrate binding by enzymes • Highly complementary interac(ons between substrate and enzyme – Hydrophobic to hydrophobic – Hydrogen bonding – Favorable Coulombic interac(ons • Substrate binding typically involves some degree of conforma(onal change in the enzyme – Enzymes need to be flexible for substrate binding and catalysis. – Provides op(mal recogni(on of substrates – Brings cataly(cally important residues to the right posi(on. • Substrate binding site is a rela(vely small region 2015, September 8 Alcohol dehydrogenase (dimer) -‐ Ethanol à acetaldehyde -‐ NADH, Zn cofactors 6 Transi(on-‐state binding vs substrate binding • Enzyme must bind substrate, transi(on state and product. • Tight binding to substrate or product slows overall reac(on by increasing the height of the barrier to TS* or product dissocia(on, respec(vely. • Tight binding to TS* speeds the reac(on. Figure 10.15 2015, September 8 PHRM 836 -‐ Devlin Ch 10 7 Transi(on states • Rate enhancement of a chemical reac(on by transi(on state stabiliza(on. • Par(al charges occur frequently in transi(on states. + Reactant Transi(on state chemical intermediate; very short-lived; high energy 2015, September 8 PHRM 836 -‐ Devlin Ch 10 Product Figure 10.13 8 Transi(on state stabiliza(on: an illustra(on Arg 127 Environment optimal for reaction! Figure 10.39, 10.40 2015, September 8 PHRM 836 -‐ Devlin Ch 10 12 Cofactors and coenzymes 1. Non-‐protein small molecules required for func(on of some enzymes 2. Organic cofactors are also called coenzymes or prosthe(c groups. – – – Many (not all) are deriva(ves of vitamins For some enzymes, are chemically modified during the reac(on Func(on: hydride or electron transfer; group transfer 3. Inorganic cofactors – – – Metal ions Metal clusters Func(on: polarize bonds; coordina(on; metal reduc(on/ oxida(on More informa+on: h.p://en.wikipedia.org/wiki/Cofactor_(biochemistry) 2015, September 8 PHRM 836 -‐ Devlin Ch 10 13 Examples of important organic cofactors or coenzymes More informa+on: h.p://en.wikipedia.org/wiki/Cofactor_(biochemistry) 2015, September 8 PHRM 836 -‐ Devlin Ch 10 Textbook of Biochemistry with Clinical Correla;ons, 7e edited by Thomas M. Devlin © 2011 John Wiley & Sons, Inc. 14 Enzyme ac(ve sites: cofactors bind as substrates zinc ethanol NAD (analogue) Alcohol dehydrogenase (dimer) -‐ Ethanol à acetaldehyde -‐ Uses zinc and NAD Ac(ve Site hjp://www.rcsb.org/pdb/101/motm_disscussed_entry.do?id=1adc#.Ujb9HgFU9us.email 2015, September 8 PHRM 836 -‐ Devlin Ch 10 15 Enzyme ac(ve sites: exquisite spa(al complementarity (i.e. stereochemistry) zinc ethanol NAD (analogue) Alcohol dehydrogenase (dimer) -‐ Ethanol à acetaldehyde -‐ Uses zinc and NAD Ac(ve Site hjp://www.rcsb.org/pdb/101/motm_disscussed_entry.do?id=1adc#.Ujb9HgFU9us.email 2015, September 8 PHRM 836 -‐ Devlin Ch 10 16 Effect of pH on enzyme catalysis • When the rate-‐limi(ng component of the cataly(c process involves a (tratable residue, the measured ac(vity of the enzyme depends on pH and the ioniza(on status of that residue. à Highest enzyma(c ac(vity occurs with the proper ioniza(on state. • Example below for general acid involvement in the cataly(c step: – Enz—AH (ac(vite) ó Enz—A-‐ (inac(ve) – Enzyme ac(vity = 50% maximum ac(vity when pH = pKa of the general acid 1.0 pKa = ? Is the proper ioniza(on state protonated or unprotonated? Relative Activity 0.8 0.6 0.4 0.2 0.0 4 5 6 7 8 9 10 pH 2015, September 8 PHRM 836 -‐ Devlin Ch 10 17 Effect of pH on enzyme catalysis Example of pH dependence due to binding • Alcohol dehydrogenase (ADH) has 3 isoforms with different pH op(ma • NADH release is the rate-‐limi(ng step ADH isoform pH Res op+ma 47 Res 369 β1 10 Arg Arg β2 8.5 His Arg β2 7.0 Arg Cys Arg 47 Arg 369 Explain the basis for this pH dependence (Clinical Correlation 10.6) 2015, September 8 PHRM 836 -‐ Devlin Ch 10 18 Summary of Enzyme Catalysis • The func(on of enzymes is in(mately linked to their structure – Specificity for substrate, cofactors (induced changes in structure) – Stabiliza(on of the transi(on state, which defines enzyme catalysis • The pH dependence of catalysis derives from interac(ons of (tratable groups formed in the rate-‐ limi(ng steps of the reac(on mechanism, whatever that step may be (bond making/breaking; product release; substrate binding, etc). 2015, September 8 PHRM 836 -‐ Devlin Ch 10 19