* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Intermolecular Interactions
Survey
Document related concepts
Transcript
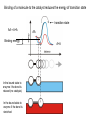
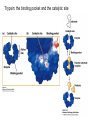
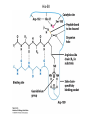
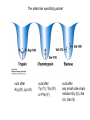
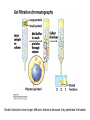
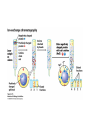
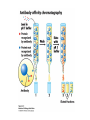
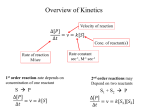
Lecture 7 Brief review of enzyme mechanisms and kinetics A ‘toolbox’ for mechanistic biochemists. A path to the molecule: protein purification techniques. Protein Kinase A spatially organizes ATP and peptide chain to facilitate the reaction Rate of ‘hopping’ over the barrier: k BT DG / k BT k e h kB – Boltzmann constant frequency pre-factor Boltzmann factor h – Plank constant DG*s→p DG*p→s DG o Binding of a molecule to the catalyst reduces the energy of transition state transition state AA = A+A Binding energy In the bound state to enzyme I the bond is relaxed (no catalysis) In the bound state to enzyme II the bond is stretched AA A+A Trypsin: the binding pocket and the catalytic site His58 Asp102 Ser195 Asp189 His58 Asp102 Ser195 Asp189 His-58 The sidechain specificity pocket cuts after Arg (R), Lys (K) cuts after Tyr (Y), Trp (W) or Phe (F) cuts after any small side-chain residue Gly (G), Ala (A), Ser (S) Catalysis involves simple binding L R LR K b [ L] B 1 K b [ L] 1 Kd Kb [ L] B K d [ L] Langmuir Michaelis-Menten equation for enzyme/transport reactions is very similar to the Langmuir isotherm V Vmax [ s] K m [ s] Vmax 1 Km = 1 mM 0.8 Km = 10 mM 0.6 0.4 0.2 0 0 10 20 30 [s], mM 40 50 A “simple explanation” says that the rate of reaction should be proportional to the occupancy of the binding site as long as Vmax is constant. Rate of ‘hopping’ over the barrier: k BT DG / k BT k e h DG*uncat E+S DG*cat E+P E + S ↔ ES ↔ EP ↔ E + P k1 k2 E + S ↔ ES → E + P (we assume k-2 = 0 ) k-1 Et – total enzyme d [ES] k1 ([ Et ] [ES])[S] (k 1[ES] k 2 [ES]) dt Rate of ES formation At steady state: d [ES] 0 dt k 1 k 2 Km k1 k 2 [Et][S] V K m [S] Rate of ES breakdown k1[Et ][S] [ES] k1[S] k 1 k 2 V k2 [ES] Vmax [S] V K m [S] Vmax k2 [Et ] Michaelis-Menten again How do we know who are the players in the structural assembly, reaction of interest or specific signaling cascade? 1. Standard biochemical ‘brute-force’ approach: isolate/purify the protein component(s) and show that they are functional or necessary for the specific function; then clone the gene(s), mutate or knock them out (down) and demonstrate altered function (biochemistry+reverse genetics). 2. Genetic approach: mutagenize and see the phenotypic deviation, find the mutant, identify and clone the gene conferring that specific trait, predict its product, generate the knock-out and show the interdependence between the phenotype and the presence of intact gene coding for that protein (forward genetics). 3. A reductionistic finale of either path toward the molecule: for most of the mechanistic studies it is important to express and isolate that protein, find the conditions under which it is active in isolation, reconstitute its functional state in a test tube and determine its properties in vitro. Obtain structural and mechanistic clues on how the molecular machine works. Cell disruption and fractionation 2D gels separate first by charge an then by size Small molecules have longer diffusion distance because they penetrate the beads Western Blotting Mass spectrometry measures m/z (mass/charge) Complete sequencing of a protein can be accomplished using MS/MS in conjunction with genomic information