* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lecture notes Chapter 22-23
Photosynthetic reaction centre wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Citric acid cycle wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Point mutation wikipedia , lookup
Western blot wikipedia , lookup
Catalytic triad wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Peptide synthesis wikipedia , lookup
Enzyme inhibitor wikipedia , lookup
Genetic code wikipedia , lookup
Metalloprotein wikipedia , lookup
Proteolysis wikipedia , lookup
Amino acid synthesis wikipedia , lookup
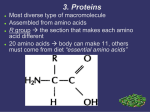
Chemistry B11 Chapters 22 & 23 Proteins and Enzymes Proteins: all proteins in humans are polymers made up from 20 different amino acids. Proteins provide structure in membranes, build cartilage, muscles, hair, nails, and connective tissue (wool, silk, feathers, and horns are some other proteins made by animals), transport oxygen in blood and muscle, direct biological reactions as enzymes, defend the body against infection, and control metabolic processes as hormones. They can even be a source of energy. Different functions of proteins depend on the structure and chemical behavior of amino acids, the building blocks of proteins. Amino acids: proteins are composed of molecular building blocks called amino acids. An amino acid contains two functional groups, an amino group (-NH2) and a carboxylic acid group (-COOH). In all of the 20 amino acids found in proteins, the amino group, the carboxylic group, and a hydrogen atom are bonded to a central carbon atom. Amino acids with this structure are called α (alpha) amino acids. Although there are many amino acids, only 20 different amino acids are present in the proteins in humans. The unique characteristics of the 20 amino acids are due to a side chain (-R), which can be an alkyl, hydroxyl, thiol, amino, sulphide, aromatic, or cyclic group. Dr. Behrang Madani Chemistry B11 Bakersfield College Zwitterion: although it is convenient to write amino acids with carboxyl (-COOH) group and amino (-NH2) group, they are usually ionized. At the pH of most body fluids, the carboxyl group loses a H+, giving –COO-, and the amino group accepts a H+ to give an ammonium ion, -NH3+. The dipolar form of an amino acid, called a Zwitterion, has a net charge of zero (it is neutral). Classification of Amino acids: 1. Nonpolar amino acids: which have alkyl or aromatic side chains, are hydrophobic (“water-fearing”). 2. Polar amino acids: which have polar side chains such as hydroxyl (-OH), thiol (-SH), and amide (-CONH2) that form hydrogen bonds with water, are hydrophilic (“water attracting”). 3. Acidic amino acids: have side chains that contain a carboxylic group (-COOH) and can ionize as a weak acid. 4. Basic amino acids: have side chains that contain an amino group that can ionize as a weak base. D and L isomer: all of the α-amino acids (except for glycine) are chiral because the α carbon is attached to four different atoms. Thus amino acids can exist as D and L isomers (enantiomers). For the L isomer the amino group, NH2, is on the left, and in the D isomer, it is on the right. In biological systems, only L amino acids are incorporated into proteins. There are D amino acids found in nature, but not in proteins. Ionization and pH: at a certain pH known as the isoelectric point (pI), the positive and negative charges are equal, which gives an overall charge of zero (no net charge). The zwitterions for polar and nonpolar amino acids typically exist at pH values of 5.0 to 6.0. However, in a solution that is more acidic than the pI (pH about 2 or 3), the –COO- group acts as a base and accepts an H+, which gives an overall positive charge to the amino acid (net charge +1): O + H3 N-CH-C-O + H3 O+ R O + H3 N-CH-C-OH + H2 O R In a solution more basic than the pI (pH from 7.6 to 10.8), the –NH3+ group acts as an acid and loses an H+, which gives the amino acid an overall negative charge (net charge –1): O + H3 N-CH-C-O + OH R Dr. Behrang Madani Chemistry B11 O H2 N-CH-C-O + H2 O R Bakersfield College As a result, the net charge on an amino acid depends on the pH of the solution in which it is dissolved. Each amino acid has a constant pI (isoelectric point) and by comparison of values of pI and pH of solution, we can find the charge of an amino acid. O + H3 N-CH-C-OH R pH 2.0 Net charge +1 OHH3 O+ O + H3 N-CH-C-O R pH 5.0 - 6.0 Net charge 0 OH+ H3 O O H2 N-CH-C-OR pH 10.0 N et ch arge -1 Peptide: the linking of two or more amino acids forms a peptide. A peptide bond is an amide bond that forms when the –COO- group of one amino acid reacts with the –NH3+ group of the next amino acid. Note: Two amino acids linked together by a peptide bond form a dipeptide, three amino acids from tripeptide, many amino acids form polypeptide. Protein is a biological macromolecule containing at least 30 to 50 amino acids joined by peptide bonds. peptide bond CH3 + H 3N O O Alanine (Ala) - + + H 3N O - O CH2 OH Serine (Ser) + H 3N CH3 H N O O - + H2 O O CH2 OH Alanylseri ne (Ala-Ser) Note: In a peptide, the amino acid written on the left with the unreacted or free amino group (-NH3+) is called the N terminal amino acid. The C terminal amino acid is the last amino acid in the chain with the unreacted or free carboxyl group (-COO-). Naming of peptide: in naming a peptide, each amino acid beginning from the N terminal is named with a “-yl” ending (we drop the “-ine” and we replace “-yl”) followed by the full name of the amino acid at the C terminal. For example, a tripeptide consisting of alanine, glycine, and serine is named as alanylglycylserine. For convenience, the order of amino acids in the peptide is often written as the sequence of three-letter abbreviations. Dr. Behrang Madani Chemistry B11 Bakersfield College Levels of protein structure: Each protein in our cells has a unique sequence of amino acids that determines in biological function. Four levels exist for structure of the proteins: 1. Primary structure 2. Secondary structure 3. Tertiary structure 4. Quaternary structure 1. Primary Structure: The primary structure is the order of the amino acids held together by peptide bonds. The first protein to have its primary structure determined was insulin, which is a hormone that regulated the glucose level in the blood. In the primary structure of human insulin, there are two polypeptide chains. In the chain A, there are 21 amino acids, and chain B has 30 amino acids. The polypeptide chains are held together by disulfide bonds formed by the side chains of the cysteine amino acids in each of the chains. Today human insulin produced through genetic engineering is used in the treatment of diabetes. Note: The -SH (sulfhydryl) group of cysteine (an amino acid) is easily oxidized to an -S-S(disulfide). + 2 H3 N-CH-COO CH2 SH Cysteine oxidation reduction + H3 N-CH-COO CH2 a disulfide bon d S S CH 2 + H3 N-CH-COO Cystine Secondary structure: the secondary structure of a protein describes the way the amino acids next to or near to each other along the polypeptide are arranged in space. The three most common types of secondary structure are the alpha helix, the beta-pleated sheet, and the triple helix found in collagen. Alpha helix (α-helix): the corkscrew shape of an alpha helix is held in place by hydrogen bonds between each N-H group and the oxygen of a C=O group in the next turn of the helix, four amino acids down the chain. Because many hydrogen bonds form along the peptide backbone, this portion of the protein takes the shape of a strong, tight coil that looks a Dr. Behrang Madani Chemistry B11 Bakersfield College telephone cord or a Slinky toy. All the side chains (R groups) of the amino acids are located on the outside of the helix. H-bond Beta-pleated sheet (-pleated sheet): in a -pleated sheet, polypeptide chains are held together side by side by hydrogen bonds between the peptide chains. In a -pleated sheet of silk fibroin, the small R groups of the prevalent amino acids, glycine, alanine, and serine, extend above and below the sheet. This result in a series of -pleated sheets that are stacked close together. The hydrogen bonds holding the -pleated sheets tightly in place account for the strength and durability of proteins such as silk. O H Triple helix (Collagen): the most abundant protein, makes up as one-third of all the protein in vertebrates. It is found in connective tissue, blood vessels, skin, tendons, ligaments, the cornea of the eye, and cartilage. The strong structure of collagen is a result of three polypeptides woven together like a braid to form a triple helix. Collagen has a high content of glycine (33%), praline (22%), alanine (12%), and smaller amounts of hydroxyproline, and hdroxylysine. The hydroxyl forms of praline and lysine contain –OH groups that form hydrogen bonds across the peptide chains and give strength to the collagen triple helix. Note: When a diet is deficient in vitamin C, collagen fibrils are weakened because the enzymes needed to form hydoxyproline and hydroxylysine require vitamin C. Without the – Dr. Behrang Madani Chemistry B11 Bakersfield College OH groups of hydroxyproline and hydroxylysine, there is less hydrogen bonding between collagen fibrils. As a person ages, additional cross-links form between the fibrils, which make collagen less elastic. Bones, cartilage, and tendons become more brittle, and wrinkles are seen as the skin loses elasticity. Tertiary structure: the tertiary structure of a protein involves attractions and repulsions between the side chain groups of the amino acids in the polypeptide chain. As interactions occur between different parts of the peptide chain, segments of the chain twist and bend until the protein acquires a specific three-dimensional shape. The tertiary structure of a protein is stabilized by interactions between the R groups of the amino acids in one region of the polypeptide chain with R groups of amino acids in other regions of the protein. There are five important interactions: 1. Hydrogen bond: occurs between H and O or N. 2. Hydrophobic interactions: are interactions between two nonpolar R groups. Within the protein, the amino acids with nonpolar side chains push as far away from the aqueous environment as possible, which forms a hydrophobic center at the interior of the molecule. 3. Hydrophilic interactions: are attractions between the external aqueous environment and amino acids that have polar or ionized side chains. The polar side chains pull toward the outer surface of globular proteins to hydrogen bond with water. 4. Salt bridges: are ionic bonds between side chains of basic and acidic amino acids, which have positive and negative charges. The attraction of the oppositely charged side chains forms a strong bond called a salt bridge. If the pH changes, the basic and acidic side chains lose their ionic charges and cannot form salt bridge, which causes a change in the shape of the protein. 5. Disulfide bonds: are the strong covalent links between sulfur atoms of two cysteine amino acids. Globular proteins: a group of proteins known as globular proteins have compact, spherical shapes because sections of the polypeptide chain fold over on top of each other. It is the globular proteins that carry out the work of the cells: functions such as synthesis, transport, Dr. Behrang Madani Chemistry B11 Bakersfield College and metabolism. Myoglobin is a globular protein that stores oxygen in skeletal muscle. High concentrations of myoglobin are found in the muscles of sea mammals, such as seals and whales, which allow them to stay under the water for long periods. Myoglobin contains 153 amino acids in a single polypeptide chain with about three-fourths of the chain in the α-helix secondary structure. Within the tertiary structure, a pocket of amino acids and a heme group binds and store oxygen. Fibrous proteins: are proteins that consist of long, thin, fiber-like shapes. They are typically involved in the structure of cells and tissue. Two types of fibrous protein are α- and keratins. The α-keratins are the proteins that make up hair, wool, skin, and nails. In hair, three α-helixes coil together like a braid to form a fibril. Within the fibril, the α-helices are held together by disulfide (-S-S-) linkages between the R groups of the many cysteine amino acids in hair. The -keratins are the type of proteins found in the feathers of birds and scales of reptiles. In -keratins, the proteins consist of large amount of -pleated sheet structure. Quaternary structure: when a biologically active protein consists of two or more polypeptide subunits, the structure level is referred to as a quaternary structure. Hemoglobin, a globular protein that transports oxygen in blood, consists of four polypeptide chains or subunits, two α chains, and two chains. The subunits are held together in the quaternary by the same interactions that stabilized the tertiary structure, such as hydrogen bonds and salt bridges between side groups, disulfide links, and hydrophobic attractions. Each subunit of the hemoglobin contains a heme group that binds oxygen. In the adult hemoglobin molecule, all four subunits must be combined for the hemoglobin to properly function as an oxygen carrier. Therefore, the complete quaternary structure of hemoglobin can bind and transport four molecules of oxygen. chain α chain chain α chain Summary of structural level in proteins: Dr. Behrang Madani Chemistry B11 Bakersfield College Denaturation of proteins: denaturation of a protein occurs when there is a disruption of any of the bonds that stabilize the secondary, tertiary, or quaternary structure. However, the covalent amide bonds of the primary structure are not affected. When the interactions between the R groups are undone or altered, a globular protein unfolds like a piece of spaghetti. With the loss of its overall shape, the protein is no longer biologically active. Denaturation agents include heat, acids and bases, organic compounds, heavy metal ions, and mechanical agitation. The following table shows some examples of proteins denaturation. Some denaturatins are reversible, while others permanently damage the protein. Enzymes: as a catalyst, an enzyme increases the rate of a reaction by changing the way a reaction takes place, but is itself not changed at the end of the reaction. An unanalyzed reaction is a cell may take place eventually, but not at a rate fast enough for survival. For example, the hydrolysis of proteins in our diet would eventually occur without a catalyst, but not fast enough to meet the body’s requirements for amino acids. The chemical reactions in our cells must occur at incredibly fast rates under the mild conditions pf pH 7.4 and a body temperature of 37°C. To do this, biological catalysts known as enzymes catalyze nearly all the chemical reactions that take place in the body. As catalysts, enzymes lower the activation energy for the reaction (activation energy is the minimum energy necessary to cause a chemical reaction to occur). As a result, less energy is required to convert reactant molecules to products, which allows more reacting molecules to form product. Dr. Behrang Madani Chemistry B11 Bakersfield College Eact Eact Names of enzymes: the names of enzymes describe the compound or the reaction that is catalyzed. The actual names of enzymes are derived by replacing the end of the name of the reaction or reacting compound with the suffix “-ase”. For example, an oxidase catalyzes an oxidation reaction, and a dehydrogenase removes hydrogen atoms. The compound amylose is hydrolyzed by the enzyme amylase, and a lipid is hydrolyzed by a lipase. Some early known enzymes use names that end in the suffix “-in”, such as papain found in papaya, rennin found in milk, and pepsin and trypsin, enzymes that catalyze the hydrolysis of proteins. There are six classes of enzymes: Enzyme action: Nearly enzymes are globular proteins. Each has a unique three-dimensional shape that recognizes and binds a small group of reacting molecules, which are called substrates. The tertiary structure of an enzyme plays an important role in how that enzyme catalyzes reactions. Active site: in a catalyzed reaction, an enzyme must first bind to a substrate in away that favors catalysis. A typical enzyme is much larger that its substrate. However, within its large tertiary structure, there is a region called the active site where the enzyme binds a substrate or substrates and catalyzes the reaction. This active site is often a small pocket that closely fits the structure of the substrate. Within the active site, the side chains of amino acids bind the Dr. Behrang Madani Chemistry B11 Bakersfield College substrate with hydrogen bonds, salt bridges, or hydrophobic attractions. The active site of a particular enzyme fits the shape of only a few types of substrates, which makes enzymes very specific about the type of substrate they bind. Enzyme catalyzed reaction: the proper alignment of a substrate within the active site forms an enzyme-substrate (ES) complex. This combination of enzyme and substrate provides an alternative pathway for the reaction that has a lower activation energy. Within the active site, the amino acid side chains take part in catalyzing the chemical reaction. As soon as the catalyzed reaction is complete, the products are quickly released from the enzyme so it can bind to a new substrate molecule. We can write the catalyzed reaction of an enzyme (E) with a substrate (S) to form product (P) as follows: E (Enzyme) + S (Substrate) ES (Complex) E (Enzyme) + P (Product) Let’s consider the hydrolysis of sucrose by sucrase (enzyme). When sucrose binds to the active site of sucrase, the glycosidic bond of sucrose is placed into a geometry favorable for reaction. The amino acid side chains catalyze the cleavage of the sucrose to give the products glucose and fructose. Because the structures of the products are no longer attracted to the active site, they are releases and the sucrase binds another sucrose substrate. Sucrase (E) + Sucrose (S) Sucrase-Sucrose complex → Sucrase (E) + glucose (P2) + Fructose (P2) There are two models for formation of ES Complex: 1. Lock-and-Key model: in this theory, the active site is described as having a rigid, nonflexible shape. Thus only those substrates with shapes that fit exactly into the active site are able to bind with enzyme. The shape of the active site is analogous to a lock, and the proper substrate is the key that fits into the lock. 2. Induced-Fit model: certain enzymes have a broader range of activity that the lock and key model allows. In the induced-fit model, enzyme structure is flexible, not rigid. There is an interaction between both the enzyme and substrate. The active site adjusts to fit the shape of the substrate more closely. At the same time the substrate adjusts its shape to better adapt to the geometry of the active site. As a result, the reacting section of the substrate becomes aligned exactly with the groups in the active site that catalyze the reaction. A different substrate could not induce these structural changes and no catalysis would occur. Dr. Behrang Madani Chemistry B11 Bakersfield College Factors affecting enzyme activity: the activity of an enzyme describes how fast an enzyme catalyzed the reaction that converts a substrate to product. This activity is strongly affected by reaction conditions, which include the temperature, pH, concentration of the substrate or enzyme and the presence of inhibitors. 1. Temperature: enzymes are very sensitive to temperature. At low temperature, most enzymes show little activity because there is not a sufficient amount of energy for the catalyzed reaction to take place. At higher temperatures, enzyme activity increases as reacting molecules move faster to cause more collisions with enzymes. Enzymes are most active at optimum temperature, which is 37°C or body temperature for most enzymes. At temperature above 50°C, the tertiary structure and thus the shape of most proteins is destroyed, which causes a loss in enzyme activity. For this reason, equipment in hospitals and laboratories is sterilized in autoclaves where the high temperatures denature the enzyme in harmful bacteria. 2. pH: enzymes are most active at their optimum pH, the pH that maintains the proper tertiary structure of the protein. A pH value above or below the optimum pH causes a change in he three-dimensional structure of the enzyme that disrupts the active site. As a result the enzyme cannot bind substrate properly and no reaction occurs. Enzymes in most cells have optimum pH values at physiological pH values around 7.4. However, enzymes in the stomach have a low optimum pH because they hydrolyze proteins at the acidic pH in the stomach. For example, pepsin, a digestive enzyme in the stomach, has an optimum pH of 2. Between meals, the pH in the stomach is 4 or 5 and pepsin shows little or no digestive activity. When food enters the stomach, the secretion of HCl lowers the pH to about 2, which actives pepsin. If small changes in pH are corrected, an enzyme can regain its structure and activity. However, large variations from optimum pH permanently destroy the structure of the enzyme. Dr. Behrang Madani Chemistry B11 Bakersfield College 3. Substrate and enzyme concentration: in any catalyzed reaction, the substrate must first bind with the enzyme to form the substrate-enzyme complex. When enzyme concentration increases, the rate of catalyzed reaction increases because we produce more substrate-enzyme complex. When enzyme concentration is kept constant, increasing the substrate concentration increases the rate of the catalyzed reaction as long as there are more enzyme molecules present than substrate molecules. At some point an increase in substrate concentration saturates the enzyme. With all the available enzyme molecules bonded to substrate, the rate of the catalyzed reaction reaches its maximum. Adding more substrate molecules cannot increase the rate further. Maximum activity 4. Enzyme inhibition: many kinds of molecules called inhibitors cause enzymes to lose catalytic activity. Although inhibitors act differently, they are all prevent the active site from binding with a substrate. Inhibition can be competitive or noncompetitive. Competitive inhibitor: a competitive inhibitor has a structure that is so similar to the substrate it competes for the active site on the enzyme. As long as the inhibitor occupies the active site, the substrate cannot bind to the enzyme and no reaction takes place. As long as the concentration of the inhibitor is substantial, there is a loss of enzyme activity. However, increasing the substrate concentration displaces more of the inhibitor molecules. As more enzyme molecules bind to substrate (ES), enzyme activity is regained. Noncompetitive inhibitor: the structure of a noncompetitive inhibitor does not resemble the substrate and does not compete for the active site. Instead, a noncompetitive inhibitor binds to a site on the enzyme that is not the active site. When the noncompetitive inhibitor is bonded to the enzyme, the shape of the enzyme is distorted. Inhibition occurs because the substrate cannot fit in the active site, or it does not fit properly. Without the proper alignment of substrate with the amino acid side groups, no catalysis can take place. Because a noncompetitive inhibitor is not competing for the active site, the addition of more substrate does not reverse this type of inhibition. Example of noncompetitive inhibitors are the heavy Dr. Behrang Madani Chemistry B11 Bakersfield College metal ions Pb2+, Ag+, and Hg2+ that bond with amino acid side groups such as –COO-, or – OH. Catalytic activity is restored when chemical reagents remove the inhibitors. Antibiotics produced by bacteria, mold, or yeast are inhibitors used to stop bacterial growth. For example, penicillin inhibits an enzyme needed for the formation of cell walls in bacteria, but not human cell membranes. With an incomplete wall, bacteria cannot survive, and the infection is stopped. However, some bacteria are resistant to penicillin because they produce penicillinase, an enzyme that breaks down penicillin. Over the years, derivatives of penicillin to which bacteria have not yet become resistant have been produced. Inhibitor Site Enzyme cofactors: enzymes are known as simple enzyme when their function forms consist only of proteins with tertiary structure. However, many enzymes require small molecules or metal ions called cofactors to catalyze reactions properly. When the cofactor is a small organic molecule, it is known as a coenzyme. If an enzyme requires a cofactor, neither the protein structure nor the cofactor alone has catalytic activity. Simple enzyme protein protein protein Metal ion Organic molecules (coenzyme) Enzyme + Cofactor Enzyme + Cofactor Metal ions: many enzymes must contain a metal ion to carry out their catalytic activity. The metal ions are bonded to one or more of the amino acid side chains. The metal ions from the minerals that we obtain from foods in our diet have various functions in catalysis. Ions such as Fe2+ and Cu2+ are used by oxidases because they lose or gain electrons in oxidation or reduction reactions. Other metals ions such as Zn2+ stabilize the amino acid side chains during hydrolysis reactions. Dr. Behrang Madani Chemistry B11 Bakersfield College Vitamins and coenzymes: vitamins are organic molecules that are essential for normal health and growth. They are required in trace amounts and must be obtained from the diet because they are not synthesized in the body. Vitamins are classified into two groups by solubility: water-soluble and fat-soluble. Water-soluble vitamins: have polar groups such as –OH and –COOH, which make them soluble in the aqueous environment of the cells. Most water-soluble enzymes are not stored in the body and excess amounts are eliminated in the urine each day. Therefore, the watersoluble vitamins must be in the foods of our daily diets. Because many water-soluble vitamins are easily destroyed by heat, oxygen, and ultraviolet light, care must be taken in food preparation, processing, and storage. The water-soluble vitamins are required by many enzymes as cofactors to carry out certain aspects of catalytic action. The coenzymes do not remain bonded to a particular enzyme, but are used over and over again by different enzymes to facilitate an enzyme-catalyzed reaction. Thus, only small amounts of coenzymes are required in the cells. Fat-soluble vitamins: are nonpolar compounds, which are soluble in the fat (lipid) components of the body such as fat deposits and cell membranes. The fat-soluble vitamins A, D, E, and K are not involved as coenzymes, but they are important in processes such as vision, formation of bone, protection from oxidation, and proper blood clotting. Because the fat-soluble vitamins are stored in the body and not eliminated, it is possible to take too much, which could be toxic. Dr. Behrang Madani Chemistry B11 Bakersfield College