* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Manolis Kellis
Secreted frizzled-related protein 1 wikipedia , lookup
Gene desert wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Gene expression profiling wikipedia , lookup
List of types of proteins wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
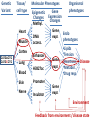
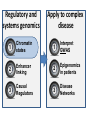
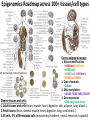
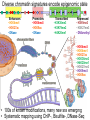
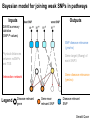
Epigenomic and regulatory genomics of complex human disease Manolis Kellis Broad Institute of MIT and Harvard MIT Computer Science & Artificial Intelligence Laboratory Family Inheritance Personal genomics today: 23 and Me Recombination breakpoints Me vs. my brother My dad Mom’s dad Disease risk Human ancestry Dad’s mom Genomics: Regions mechanisms drugs AMD Risk Systems: genes combinations pathways Genetic Variant Tissue/ cell type Heart Muscle Cortex CATGACTG CATGCCTG Lung Blood Skin Nerve Molecular Phenotypes Organismal phenotypes Gene Epigenetic Expression Changes Changes Methyl. Gene Endo DNA expr. phenotypes access. Lipids Tension Enhancer Gene Heartrate Disease expr. Metabol. H3K27ac Drug resp Promoter Gene expr. Insulator Environment Feedback from environment / disease state Regulatory and systems genomics Apply to complex disease 1 Chromatin states 1 Interpret GWAS 2 Enhancer linking 2 Epigenomics in patients 3 Causal Regulators 3 Disease Networks Epigenomics Roadmap across 100+ tissues/cell types Art: Rae Senarighi, Richard Sandstrom Diverse epigenomic assays: 1. Histone modifications • H3K4me3, H3K4me1 • H3K36me3 • H3K27me3, H3K9me3 • H3K27ac, H3K9ac 2. Open chromatin: • DNase 3. DNA methylation: • WGBS, RRBS, MRE/MeDIP 4. Gene expression • RNA-seq, Exon Arrays Diverse tissues and cells: 1. Adult tissues and cells (brain, muscle, heart, digestive, skin, adipose, lung, blood…) 2. Fetal tissues (brain, skeletal muscle, heart, digestive, lung, cord blood…) 3. ES cells, iPS, differentiated cells (meso/endo/ectoderm, neural, mesench, trophobl) Diverse chromatin signatures encode epigenomic state Enhancers • H3K4me1 • H3K27ac • DNase Promoters • H3K4me3 • H3K9ac • DNase Transcribed • H3K36me3 • H3K79me2 • H4K20me1 Repressed • H3K9me3 • H3K27me3 • DNAmethyl • • • • • • • • H3K4me3 H3K4me1 H3K27ac H3K36me3 H4K20me1 H3K27me3 H3K9me3 H3K9ac • 100s of known modifications, many new still emerging • Systematic mapping using ChIP-, Bisulfite-, DNase-Seq Deep sampling of 9 reference epigenomes (e.g. IMR90) UWash Epigenome Browser, Ting Wang Chromatin state+RNA+DNAse+28 histone marks+WGBS+Hi-C Chromatin states capture combinations and dynamics Predicted linking • • • • Correlated activity Single annotation track for each cell type Capture combinations of histone marks Summarize cell-type activity at a glance Study activity pattern across cell types Chromatin state annotations across 127 epigenomes Reveal epigenomic variability: enh/prom/tx/repr/het Anshul Kundaje 2.3M enhancer regions only ~200 activity patterns dev/morph immune muscle morph learning Wouter Meuleman <3 smooth muscle kidney liver Systematic motif dissection in 2000 enhancers: 5 activators and 2 repressors in 2 cell lines 54000+ measurements (x2 cells, 2x repl) Kheradpour et al Genome Research 2013 Example activator: conserved HNF4 motif match WT expression specific to HepG2 Motif match disruptions reduce expression to background Non-disruptive changes maintain expression Random changes depend on effect to motif match Regulatory and systems genomics Apply to complex disease 1 Chromatin states 1 Interpret GWAS 2 Enhancer linking 2 Epigenomics in patients 3 Causal Regulators 3 Disease Networks The challenge of interpreting disease-association studies • Large associated blocks with many variants: Fine-mapping challenge • No information on cell type/mechanism, most variants non-coding Epigenomic annotations help find relevant cell types / nucleotides Revisiting diseaseassociated variants xx • Disease-associated SNPs enriched for enhancers in relevant cell types • E.g. lupus SNP in GM enhancer disrupts Ets1 predicted activator Mechanistic predictions for top disease-associated SNPs Lupus erythromatosus in GM lymphoblastoid Erythrocyte phenotypes in K562 leukemia cells ` Disrupt activator Ets-1 motif Loss of GM-specific activation Loss of enhancer function Loss of HLA-DRB1 expression Creation of repressor Gfi1 motif Gain K562-specific repression Loss of enhancer function Loss of CCDC162 expression GWAS hits in enhancers of relevant cell types Immune traits, heart, height, platelets, in relevant tissues Luke Ward Rank-based functional testing of weak associations Enrichment peaks at 10,000s of SNPs down the rank list, even after LD pruning! Abhishek Sarkar • Rank all SNPs based on GWAS signal strength • Functional enrichment for cell types and states Weak-effect T1D hits in 1000s T-cell enhancers enhancers CD4+ T-cells T-cells B-cells Other cell types Abhishek Sarkar • Enhancer enrichment strong for top ~30k SNPs • Heritability estimates also increase until ~30k SNPs Brain methylation changes in AD patients Per state: (Obs – Exp) / Total Enhancers Promoters • 10,000s of methylation differences in AD vs. control • Harbor 1000s of genetic variants associated with AD • Localized in brain-specific enhancers and pathways T1D/RA-enriched enhancers spread across genome Abhishek Sarkar • High concentration of loci in MHC, high overlap • Yet: many distinct regions, 1000s of distinct loci Bayesian model for joining weak SNPs in pathways Inputs Outputs GWAS summary statistics (SNP P-values) SNP disease-relevance (yes/no) Physical distances between ncSNPs and TSS Gene target (if any) of each SNP3 Interaction network Gene disease-relevance (yes/no) Legend Disease-relevant gene Gene near relevant SNP Disease-relevant SNP Gerald Quon Highly ranked SNP nearby 200 400 600 800 Poorly ranked SNP nearby 0 0 # SNPs whose p>0 # SNPs (p>0) 1200 1200 Example 1: MAZ predicted role in T1D 00.0 0.2 0.4 0.6 0.8 p(SNP relevant) 11.0 p(SNP is disease−relevant) 10000 5000 # genes # genes 15000 15k 0 0 0 0.0 0.2 0.4 0.6 0.8 1 1.0 p(gene relevant) p(gene is disease−relevant) Gerald Quon • MAZ no direct assoc, but clusters w/ many T1D hits • MAZ indeed known regulator of insulin expression Example 2: SP3 predicted role in MS 300 Highly ranked SNP nearby 50 100 200 Poorly ranked SNP nearby 0 0 # SNPs whose p>0 # SNPs (p>0) 300 0 0.0 0.2 0.4 0.6 0.8 p(SNP relevant) 1 1.0 p(SNP is disease−relevant) 6000 4000 2000 # genes # genes 8000 8k 0 0 0 0.0 0.2 0.4 0.6 0.8 p(gene relevant) p(gene is disease−relevant) 1 1.0 Gerald Quon • SP3 no direct assoc but clusters w/ many MS hits • SP3 is indeed down-regulated in MS patients # non-genetic hits missing heritability • Missing heritability partly due to weak variants • Regulators lacking association harbor rare variants e.g. Coronary artery disease: GATA6 (congential heart disease), HNF1A (cardiovascular), PPARG (lipid metabolism, partial lipodystrophy) Gerald Quon Validate weak variant targets in model organisms Use CRISPR/Cas to edit nucleotides, knockdown target genes Alzheimer: Differential activity in mouse neurodegeneration Andreas Pfenning Cardiac: Repolarization interval in zebrafish heart Xinchen Wang Regulatory and systems genomics Apply to complex disease 1 Chromatin states 1 Interpret GWAS 2 Enhancer linking 2 Epigenomics in patients 3 Causal Regulators 3 Disease Networks Integrative analysis of 100+ epigenomes 1. Reference Epigenomes chromatin states, linking – Annotate dynamic regulatory elements in multiple cell types – Activity-based linking of regulators enhancers targets 2. Interpreting disease-associated sequence variants – Mechanistic predictions for individual top-scoring SNPs – Functional roles of 1000s of disease-associated SNPs 3. Disease networks: links SNPsgenesphenotypes – Module-based linking of enhancers to their target genes – Bayesian model for evaluating disease genes and SNPs 4. Genetic / epigenomic variation in health and disease – Genetic variationBrain methylationAlzheimer’s disease – Global repression of distal enhancers. NRSF, ELK1, CTCF MIT Computational Biology Group Hayden Metsky Anshul Andreas Matt Luis Abhishek Stefan Kundaje Pfenning Eaton Barrera Sarkar Washietl Bob Altshuler Manasi Vartak Daniel Marbach Jessica Wu Pouya David Mariana Kheradpour Hendrix Mendoza Matt Rasmussen Manolis Mukul Kellis Bansal Wouter Meuleman Gerald Quon Soheil Feizi Jason Ernst Luke Ward Roadmap Epigenomics Integrative Analysis Team Anshul Kundaje Wouter Meuleman Jason Ernst Misha Bilenky Lisa Chadwick Jianrong Wang Ting Wang Angela Yen John Stam Luke Ward Bing Ren Cristian Coarfa, Alan Harris, Michael Ziller, Matthew Abhishek Sarkar Martin Hirst Schultz, Matt Eaton, Andreas Pfenning, Xinchen Wang, Gerald Quon Joe Costello Paz Polak, Rosa Karlic, Viren Amin, Yi-Chieh Wu, Pouya Kheradpour Brad Bernstein Richard S Sandstrom, Zhizhuo Zhang, Alireza Heravi-Moussavi GiNell Elliott, Rebecca Lowdon Aleks Milosavljevic