* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Prescott`s Microbiology, 9th Edition Chapter 29 –Methods in
Point mutation wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Molecular Inversion Probe wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Genome (book) wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Microevolution wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Metagenomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Gene expression profiling wikipedia , lookup
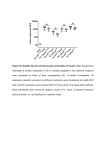
Prescott’s Microbiology, 9th Edition Chapter 29 –Methods in Microbial Ecology GUIDELINES FOR ANSWERING THE MICRO INQUIRY QUESTIONS Figure 29.4 What happens to the flow stream in fluorescence activated cell sorting (FACS)? The droplets have a net electrical charge on them, and thus have their flight directed by electric fields via attraction or repulsion. Thus, FACS will sort and collect droplets into different bins, based on set fluorescence parameters. Figure 29.7 Under what circumstances would one use epifluorescence microscopy, rather than flow cytometry, and vice versa? Epifluoresence allows you to directly visualize the cells, and see where the fluorescence is inside the cell. Flow can count large numbers of cells is a short period of time, but you don’t directly visualize them. Figure 29.8 How many different phylotypes appear to be represented by this DGGE? Each sample appears to have three different sequences. Figure 29.9 How do phylochip differ from the microarrays discussed in chapter 18 (p. 434)? In technical details, both microarrays are the same. With the phylochip application, the probes on the chip match 16S sequences unique to many different species. With the expression microarray, the probes match different gene sequences so the expression of many different genes can be simultaneously determined. 1 © 2014 by McGraw-Hill Education. This is proprietary material solely for authorized instructor use. Not authorized for sale or distribution in any manner. This document may not be copied, scanned, duplicated, forwarded, distributed, or posted on a website, in whole or part.