Lectures 21, 22, and 23: Phylogenic Trees and Evolution Steven
... • Each internal node is labeled by a number. • Each internal node has at least two children • Along any root to leaf path the labels strictly decrease. Note that if the labels mean “time units ago”, this is true of any evolutionary tree. ...
... • Each internal node is labeled by a number. • Each internal node has at least two children • Along any root to leaf path the labels strictly decrease. Note that if the labels mean “time units ago”, this is true of any evolutionary tree. ...
Chapter 26 Presentation-Phylogeny and the Tree of Life
... For example, let’s look at hinged jaws. These are absent in lampreys, but are found in other members of the ingroup-this represents a branch point. The cladogram we’ve developed isn’t a phylogenetic tree, we need more information from fossils, etc. to indicate when the groups first appeared. ...
... For example, let’s look at hinged jaws. These are absent in lampreys, but are found in other members of the ingroup-this represents a branch point. The cladogram we’ve developed isn’t a phylogenetic tree, we need more information from fossils, etc. to indicate when the groups first appeared. ...
Lecture #1: Phylogeny & the “Tree of Life”
... “twigs” to phylogenetic trees • keep in mind - different genes evolve at different rates – the evolution of ribosomal genes are slow – allows us to investigate further back in time – the evolution of mitochondrial genes are fast – investigation of more recent ...
... “twigs” to phylogenetic trees • keep in mind - different genes evolve at different rates – the evolution of ribosomal genes are slow – allows us to investigate further back in time – the evolution of mitochondrial genes are fast – investigation of more recent ...
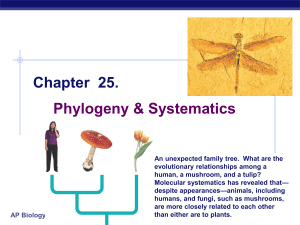
Ch_25 Phylogeny and Systematics
... apply principle of parsimony simplest explanation fewest evolutionary events that explain data ...
... apply principle of parsimony simplest explanation fewest evolutionary events that explain data ...
Bio 1B, Spring, 2007, Evolution section 1 of 4 Updated 3/21/07 7:49
... • In a phylogenetic classification, species in the same genus had a most recent common ancestor more recently than species in different genera in the same family, and so on. • Phylogenetic systematics is based on the cladogram, not on the phylogenetic tree. Finding the right cladogram The problem ...
... • In a phylogenetic classification, species in the same genus had a most recent common ancestor more recently than species in different genera in the same family, and so on. • Phylogenetic systematics is based on the cladogram, not on the phylogenetic tree. Finding the right cladogram The problem ...
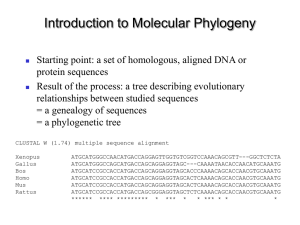
introduction to molecular phylogeny
... ATGCATCCGCCACCATGACCAGCAGGAGGTAGCactCAAAACAGCACCaacGTGCAAATG ATGCATCCGCCACCATGACCAGCGGGAGGTAGCtctCAAAACAGCACCaacGTGCAAATG ...
... ATGCATCCGCCACCATGACCAGCAGGAGGTAGCactCAAAACAGCACCaacGTGCAAATG ATGCATCCGCCACCATGACCAGCGGGAGGTAGCtctCAAAACAGCACCaacGTGCAAATG ...
Diapositiva 1 - Universidad Autonoma de Madrid
... Homoplasy in molecular data • Incongruence and therefore homoplasy can be common in molecular sequence data • One reason is that characters have a limited number of alternative character states ( e.g. A, G, C and T) • In addition, these states are chemically identical so that homology and homoplas ...
... Homoplasy in molecular data • Incongruence and therefore homoplasy can be common in molecular sequence data • One reason is that characters have a limited number of alternative character states ( e.g. A, G, C and T) • In addition, these states are chemically identical so that homology and homoplas ...
In the article entitled ‘Search for a Tree of Life... evolution, at least as far as bacteria and archaea are
... and edges reflecting gene exchange [18]. The stakes here are high because replacement of the TOL with a network graph would change our entire perception of the process of evolution and invalidate all evolutionary reconstruc tion based on a species tree. However, the tree repre sentation is by no ...
... and edges reflecting gene exchange [18]. The stakes here are high because replacement of the TOL with a network graph would change our entire perception of the process of evolution and invalidate all evolutionary reconstruc tion based on a species tree. However, the tree repre sentation is by no ...
PPTX - Tandy Warnow
... alignments and trees using treelength criteria (however – note that the developers of POY say to ignore the alignment and only use the tree). • BeeTLe (Better Tree Length) is a heuristic that is guaranteed to always be as least as accurate as POY for the treelength criterion. • The accuracy of the f ...
... alignments and trees using treelength criteria (however – note that the developers of POY say to ignore the alignment and only use the tree). • BeeTLe (Better Tree Length) is a heuristic that is guaranteed to always be as least as accurate as POY for the treelength criterion. • The accuracy of the f ...
Talk in Powerpoint Format
... • Objective: determine probability estimates that a given sample belongs to a class Probability(x Class | attribute values) • Baysian network: – One node for each attribute – Nodes connected in an acyclic graph – Conditional independance ...
... • Objective: determine probability estimates that a given sample belongs to a class Probability(x Class | attribute values) • Baysian network: – One node for each attribute – Nodes connected in an acyclic graph – Conditional independance ...
Chapter 2 - FacStaff Home Page for CBU
... 'parallel' evolution is a false dichotomy, at best representing ends of a continuum. We can simplify our vocabulary; all instances of the independent evolution of a given phenotype can be described with a single term - convergent." "If the use of the terms 'parallelism' and 'convergence' cannot be a ...
... 'parallel' evolution is a false dichotomy, at best representing ends of a continuum. We can simplify our vocabulary; all instances of the independent evolution of a given phenotype can be described with a single term - convergent." "If the use of the terms 'parallelism' and 'convergence' cannot be a ...
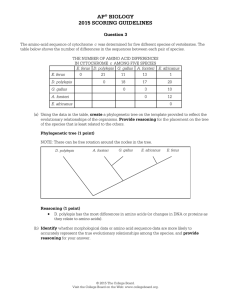
ap® biology 2015 scoring guidelines
... This question focused on using evidence to support biological evolution. Students were asked to evaluate amino acid sequences from five related species to construct a phylogenetic tree reflecting the evolutionary relationships among them, and justify the placement on the tree of the species that is ...
... This question focused on using evidence to support biological evolution. Students were asked to evaluate amino acid sequences from five related species to construct a phylogenetic tree reflecting the evolutionary relationships among them, and justify the placement on the tree of the species that is ...
TAXONOMY and EVOLUTION Test Review: Complete the following
... 3. What is the name of our scientific name system? 4. What are the levels of classification in order? 5. How are scientific names written? 6. Using a scientific name, how can you tell if two organisms are closely related? 7. What are the three domains and the 6 kingdoms that belong to them? 8. Defin ...
... 3. What is the name of our scientific name system? 4. What are the levels of classification in order? 5. How are scientific names written? 6. Using a scientific name, how can you tell if two organisms are closely related? 7. What are the three domains and the 6 kingdoms that belong to them? 8. Defin ...
Branching Activity Instructions:
... In what ways does this activity model an actual evolutionary branching tree? Hint: In what ways is this activity similar to a real situation? ________________________________________________________________________ ________________________________________________________________________ In what ways ...
... In what ways does this activity model an actual evolutionary branching tree? Hint: In what ways is this activity similar to a real situation? ________________________________________________________________________ ________________________________________________________________________ In what ways ...
PHYOGENY & THE Tree of life
... search for trees that are parsimonious & have a high probability ...
... search for trees that are parsimonious & have a high probability ...
Slide 1
... annotation of a genomic sequence • Determine ability to correlate genes to the particular phenotype • Determine ability to use BLAST to obtain orthologous sequences • Explain how genes diverge at the molecular level through the process of evolution • Determine students’ confidence in ability to cons ...
... annotation of a genomic sequence • Determine ability to correlate genes to the particular phenotype • Determine ability to use BLAST to obtain orthologous sequences • Explain how genes diverge at the molecular level through the process of evolution • Determine students’ confidence in ability to cons ...
Proposal to change linear sequence of orders to place Galliformes
... al. (2001) found that Passeriformes were basal to all Neognaths or even all living birds. These studies can be faulted, as the authors themselves often pointed out, for combinations of limited taxon sampling, rooting the tree with distantly related alligator sequence, or assuming equal rates of mtDN ...
... al. (2001) found that Passeriformes were basal to all Neognaths or even all living birds. These studies can be faulted, as the authors themselves often pointed out, for combinations of limited taxon sampling, rooting the tree with distantly related alligator sequence, or assuming equal rates of mtDN ...
Studying the evolution of photosynthesis using phylogenetic trees
... ancestors [2]. Mapping the evolutionary relationships of all living beings have been undertaken by comparing highly conserved gene sequences present in all of the above [1] and considerable intellectual effort has been invested to reduce the systematic errors of sequence alignment [1] and tree const ...
... ancestors [2]. Mapping the evolutionary relationships of all living beings have been undertaken by comparing highly conserved gene sequences present in all of the above [1] and considerable intellectual effort has been invested to reduce the systematic errors of sequence alignment [1] and tree const ...
Molecular evolution and phylogenetic implications in clinical research
... UPGMA is one of the simplest methods of phylogenetic trees construction. It has numerous limitations and wrong assumptions; therefore, this method is not readily used. The UPGMA algorithm assumes that the tree is additive (the distance between any two nodes is equal to the total length of the branch ...
... UPGMA is one of the simplest methods of phylogenetic trees construction. It has numerous limitations and wrong assumptions; therefore, this method is not readily used. The UPGMA algorithm assumes that the tree is additive (the distance between any two nodes is equal to the total length of the branch ...
Computational Biology
... Maximum likelihood approach Method uses probability calculations to find a tree that best accounts for the variation in a set of sequences. Similar to maximum parsimony method in that analysis is performed on each column of a multiple sequence alignment. All trees are considered. Because the rate o ...
... Maximum likelihood approach Method uses probability calculations to find a tree that best accounts for the variation in a set of sequences. Similar to maximum parsimony method in that analysis is performed on each column of a multiple sequence alignment. All trees are considered. Because the rate o ...
File
... The main idea of decision tree construction tree is to evaluate different attributes and different partitioning conditions, and pick the attributes and partitioning condition that results in the maximum information gain ratio. The same procedure works recursively on each of the sets resulting from t ...
... The main idea of decision tree construction tree is to evaluate different attributes and different partitioning conditions, and pick the attributes and partitioning condition that results in the maximum information gain ratio. The same procedure works recursively on each of the sets resulting from t ...
CSCE590/822 Data Mining Principles and Applications
... Rooting a tree, and definition of outgroup Neighbor-joining produces an unrooted tree How do we root a tree between N species using n-j? An outgroup is a species that we know to be more distantly related to all remaining species, than they are to one another Example: Human, mouse, rat, pig, dog, ch ...
... Rooting a tree, and definition of outgroup Neighbor-joining produces an unrooted tree How do we root a tree between N species using n-j? An outgroup is a species that we know to be more distantly related to all remaining species, than they are to one another Example: Human, mouse, rat, pig, dog, ch ...
Natural selection and phylogenetic analysis
... extent that phylogenetic analysis groups them together (6). However, long branch attraction is predicted to yield a pattern in which the sites with the highest evolutionary rate show the greatest signal favoring the wrong tree (7); this was not the case in their data. A careful process of eliminati ...
... extent that phylogenetic analysis groups them together (6). However, long branch attraction is predicted to yield a pattern in which the sites with the highest evolutionary rate show the greatest signal favoring the wrong tree (7); this was not the case in their data. A careful process of eliminati ...
Analysis of Crop Plant Genomes
... represented as a tree of evolution (often called a phylogenetic tree). ...
... represented as a tree of evolution (often called a phylogenetic tree). ...
Phylogenetic Trees - Elhanan Borenstein
... each sequence to a list of N tree nodes. 2) look through current list of nodes (initially these are all leaf nodes) for the pair with the smallest distance. 3) merge the closest pair, remove the pair of nodes from the list and add the merged node to the list. ...
... each sequence to a list of N tree nodes. 2) look through current list of nodes (initially these are all leaf nodes) for the pair with the smallest distance. 3) merge the closest pair, remove the pair of nodes from the list and add the merged node to the list. ...