A Phase Transition in Molecular Evolution with Applications to the Assembly of the Tree of Life
... Recent advances in DNA sequencing technology have led to new challenges in the analysis of the massive data sets produced in current evolutionary studies. In particular, the estimation of evolutionary trees on increasingly large scales has prompted the development of novel reconstruction methods. An ...
... Recent advances in DNA sequencing technology have led to new challenges in the analysis of the massive data sets produced in current evolutionary studies. In particular, the estimation of evolutionary trees on increasingly large scales has prompted the development of novel reconstruction methods. An ...
Document
... • The 3/4 and 4/3 terms reflect that there are four types of nucleotides and three ways in which a second nucleotide may not match a first - with all types of change being equally likely (i.e. unrelated sequences should be 25% identical by chance alone) ...
... • The 3/4 and 4/3 terms reflect that there are four types of nucleotides and three ways in which a second nucleotide may not match a first - with all types of change being equally likely (i.e. unrelated sequences should be 25% identical by chance alone) ...
CSCE590/822 Data Mining Principles and Applications
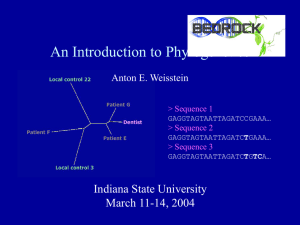
... So what do the results mean? • 2 of 3 patients closer to dentist than to local controls. Statistical significance? More powerful analyses? • Do we have enough data to be confident in our conclusions? What additional data would help? • If we determine that the dentist’s virus is linked to those of p ...
... So what do the results mean? • 2 of 3 patients closer to dentist than to local controls. Statistical significance? More powerful analyses? • Do we have enough data to be confident in our conclusions? What additional data would help? • If we determine that the dentist’s virus is linked to those of p ...
Shared character
... 1. choose organisms—they go in left column, and put the outgroup there too. Top, going right, characteristics they could share 2. score the organisms lacking certain characteristic 0, and if it has it, score 1 3. most commonly shared derived character (this case vascular tissues) goes at the base of ...
... 1. choose organisms—they go in left column, and put the outgroup there too. Top, going right, characteristics they could share 2. score the organisms lacking certain characteristic 0, and if it has it, score 1 3. most commonly shared derived character (this case vascular tissues) goes at the base of ...
lecture 03 - phylogenetics - Cal State LA
... - how were ancestral traits modified in different lineages? A hypothesis of the evolutionary history of a group is called its phylogeny - often summarized in a branching diagram called a phylogenetic tree Since we can’t travel back in time to identify common ancestors, relationships of existing spec ...
... - how were ancestral traits modified in different lineages? A hypothesis of the evolutionary history of a group is called its phylogeny - often summarized in a branching diagram called a phylogenetic tree Since we can’t travel back in time to identify common ancestors, relationships of existing spec ...
Lecture 7-POSTED-BISC441-2012
... for finding trees with the best value for the objective function. Can identify many equally optimal trees, if such exist. Warning: Finding an optimal tree is not necessarily the same as finding the "true” tree. Random data will give you an ‘optimal’ (best ) tree! ...
... for finding trees with the best value for the objective function. Can identify many equally optimal trees, if such exist. Warning: Finding an optimal tree is not necessarily the same as finding the "true” tree. Random data will give you an ‘optimal’ (best ) tree! ...
An Introduction to Phylogenetics
... more taxa that are known to have diverged prior to the group being studied • The node where the outgroup lineage joins the other taxa is the root ...
... more taxa that are known to have diverged prior to the group being studied • The node where the outgroup lineage joins the other taxa is the root ...
Evolution, 2e
... Genes for 11 tRNAs 6 proteins Human-chimpanzee relationship 1023 more likely than Chimpanzee-gorilla relationship ...
... Genes for 11 tRNAs 6 proteins Human-chimpanzee relationship 1023 more likely than Chimpanzee-gorilla relationship ...
Phylogenetics Molecular Phylogenetics
... a problem because of some change in behaviour? Or is there another explanation for their origin? Host species tree ...
... a problem because of some change in behaviour? Or is there another explanation for their origin? Host species tree ...
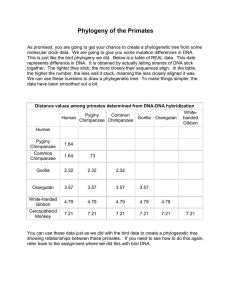
Phylogeny of the Primates
... As promised, you are going to get your chance to create a phylogenetic tree from some molecular clock data. We are going to give you some mutation differences in DNA. This is just like the bird phylogeny we did. Below is a table of REAL data. This date represents difference in DNA. It is obtained by ...
... As promised, you are going to get your chance to create a phylogenetic tree from some molecular clock data. We are going to give you some mutation differences in DNA. This is just like the bird phylogeny we did. Below is a table of REAL data. This date represents difference in DNA. It is obtained by ...
Trees
... Amino-acid sites are partially ordered characters. An amino acid cannot change into all other amino acids in a singe step, as sometimes 2 or 3 steps are required. For example, a tyrosine may only change into a leucine through an intermediate state, i.e., phenylalanine or histidine. ...
... Amino-acid sites are partially ordered characters. An amino acid cannot change into all other amino acids in a singe step, as sometimes 2 or 3 steps are required. For example, a tyrosine may only change into a leucine through an intermediate state, i.e., phenylalanine or histidine. ...
Classification and phylogeny – Chapter 2
... (Cercopithecidae) and Homo – Oldest fossils of Cercopithidae are dated at 25 mya • Average rate for Rhesus monkey lineage = 457/25 my = 1.83 x 10-3 per my • Average rate for Homo line is 310/25 my = 1.24 x 10-3 per my • Average rate is 1.54 x 10-3 per my ...
... (Cercopithecidae) and Homo – Oldest fossils of Cercopithidae are dated at 25 mya • Average rate for Rhesus monkey lineage = 457/25 my = 1.83 x 10-3 per my • Average rate for Homo line is 310/25 my = 1.24 x 10-3 per my • Average rate is 1.54 x 10-3 per my ...
Diagrams Nov 8
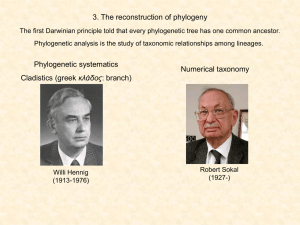
... Phenetic: classification of the organisms based on their similarities, trees obtained using a phenetic approach may not reflect evolutionary relationships. A tree based on this method is called a phenogram Cladistic (Hennig 1966): study of the different pathways of evolution, the most parsimonious p ...
... Phenetic: classification of the organisms based on their similarities, trees obtained using a phenetic approach may not reflect evolutionary relationships. A tree based on this method is called a phenogram Cladistic (Hennig 1966): study of the different pathways of evolution, the most parsimonious p ...
CIPRES.2006.algorthms_sr
... • Breakthrough: Optimal logarithmic sequence length tree reconstruction (Daskalakis, Mossel, Roch 05). Simplified version (Mihaescu et al. 06). Preliminary Implementation [Adkins et al.]. ...
... • Breakthrough: Optimal logarithmic sequence length tree reconstruction (Daskalakis, Mossel, Roch 05). Simplified version (Mihaescu et al. 06). Preliminary Implementation [Adkins et al.]. ...
Slajd 1
... The construction of phylogenetic trees from numerical methods The principle of maximum parsimony (Occam’s razor) holds that we should accept that phylogenetic tree that can be constructed with the least number of morphological changes. The raw data Species A B C D E ...
... The construction of phylogenetic trees from numerical methods The principle of maximum parsimony (Occam’s razor) holds that we should accept that phylogenetic tree that can be constructed with the least number of morphological changes. The raw data Species A B C D E ...
Comparative Anatomy: Phylogenetics Assignment
... 4. Print out a distance matrix in which you include all characters in the calculations. From looking at the distances, answer the following questions: a. Is there any reason to think that the dataset you are using will not be useful in determining phylogenetic relationships among taxa? Write your an ...
... 4. Print out a distance matrix in which you include all characters in the calculations. From looking at the distances, answer the following questions: a. Is there any reason to think that the dataset you are using will not be useful in determining phylogenetic relationships among taxa? Write your an ...
Using HIV Data Sets for Inquiry
... • Minimizes distance between nearest neighbors Maximum parsimony • Minimizes total evolutionary change Maximum likelihood • Maximizes likelihood of observed data ...
... • Minimizes distance between nearest neighbors Maximum parsimony • Minimizes total evolutionary change Maximum likelihood • Maximizes likelihood of observed data ...
Lecture5
... Creates noise in the data Some characters give conflicting information about relationships Systematists try to minimize homoplasy in a data set Choose characters that evolve slowly relative to age of taxa ...
... Creates noise in the data Some characters give conflicting information about relationships Systematists try to minimize homoplasy in a data set Choose characters that evolve slowly relative to age of taxa ...
Day6
... time, this process may repeat itself, so that at any time, each population can be said to be most closelyrelated to some other population with which it shares a direct common ancestor. ...
... time, this process may repeat itself, so that at any time, each population can be said to be most closelyrelated to some other population with which it shares a direct common ancestor. ...
Diapositiva 1
... GENE PHYLOGENIES • Parsimony (Fitch 1977), • Pairwise distances (Saitou and Nei 1987), • Maximum likelihood (Felsenstein 1981): Use of explicit models of sequence evolution (computationally very intensive) Divergence dates of genes and species can also be estimated from phylogenetic distances (Ramb ...
... GENE PHYLOGENIES • Parsimony (Fitch 1977), • Pairwise distances (Saitou and Nei 1987), • Maximum likelihood (Felsenstein 1981): Use of explicit models of sequence evolution (computationally very intensive) Divergence dates of genes and species can also be estimated from phylogenetic distances (Ramb ...
Introduction to Phylogenetics - Lectures For UG-5
... Making trees using character-based methods The main idea of character based methods is to search for a tree that requires the smallest number of evolutionary changes to explain the differences among the OTUs under study. ...
... Making trees using character-based methods The main idea of character based methods is to search for a tree that requires the smallest number of evolutionary changes to explain the differences among the OTUs under study. ...
Creating Phylogenetic Trees with MEGA
... Kumar, S., Dudley, J., Nei, M., and Tamura, K., MEGA: A biologist‐centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics 9: 299‐306 (2008) Hulsenbeck, J.P., and Ronquist, F., MyBayes: Bayesian inferences of phylogeny. Bioinformatics ...
... Kumar, S., Dudley, J., Nei, M., and Tamura, K., MEGA: A biologist‐centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics 9: 299‐306 (2008) Hulsenbeck, J.P., and Ronquist, F., MyBayes: Bayesian inferences of phylogeny. Bioinformatics ...
m12-comparative_genomics
... the one that explains observed changes using the smallest number of point mutations o Maximum Likelihood: Analog of Maximum Parsimony that attempts to identify the most likely tree rather than the cheapest one; these are the most common methods used today A single-gene tree does not always reflect ...
... the one that explains observed changes using the smallest number of point mutations o Maximum Likelihood: Analog of Maximum Parsimony that attempts to identify the most likely tree rather than the cheapest one; these are the most common methods used today A single-gene tree does not always reflect ...
Phylogenetic - Nematode bioinformatics. Analysis tools and data
... divergence order of taxa, as well as the lengths of the branches that connect them. There are many phylogenetic methods available today, each having strengths and weaknesses. Most can be classified as follows: ...
... divergence order of taxa, as well as the lengths of the branches that connect them. There are many phylogenetic methods available today, each having strengths and weaknesses. Most can be classified as follows: ...