20 years and 22 papers with Bernard Moret
... • The state at the root is randomly drawn from {A,C,T,G} (nucleotides) • If a site (position) changes on an edge, it changes with equal probability to each of the remaining states. • The evolutionary process is Markovian. The different sites are assumed to evolve independently and identically down t ...
... • The state at the root is randomly drawn from {A,C,T,G} (nucleotides) • If a site (position) changes on an edge, it changes with equal probability to each of the remaining states. • The evolutionary process is Markovian. The different sites are assumed to evolve independently and identically down t ...
Lectures 11 Friday, October 22, 2010 Phylogenetic tree (phylogeny
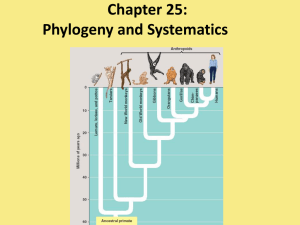
... Phylogenetic systematics: The overall goal is to make higher categories reflect evolutionary history by requiring that all taxa be monophyletic. In a phylogenetic classification, species in the same genus had a most recent common ancestor more recently than species in different genera in the same fa ...
... Phylogenetic systematics: The overall goal is to make higher categories reflect evolutionary history by requiring that all taxa be monophyletic. In a phylogenetic classification, species in the same genus had a most recent common ancestor more recently than species in different genera in the same fa ...
Lecture 4: (Part 1) Phylogenetic inference
... 2. Tree should use characters that are shared (among two or more taxa) and derived (from some inferred or known ancestral state). • shared and derived characters are called synapomorphies. 3. Ancestral state of characters inferred from an outgroup that roots the tree. • an outgroup is ideally picked ...
... 2. Tree should use characters that are shared (among two or more taxa) and derived (from some inferred or known ancestral state). • shared and derived characters are called synapomorphies. 3. Ancestral state of characters inferred from an outgroup that roots the tree. • an outgroup is ideally picked ...
PHYLOGENETIC NETWORKS
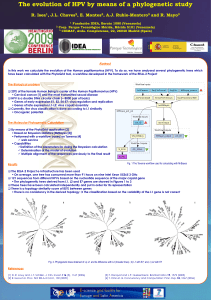
... Solving the Systematic Challenge: Our Method SpNet Given the sequences of two genes I & II on a set of species • Run MP or ML on gene I and obtain a set U1 of trees, represented by its consensus tree t1 • Run MP or ML on gene II and obtain a set U2 of trees, represented by its consensus tree t2 • F ...
... Solving the Systematic Challenge: Our Method SpNet Given the sequences of two genes I & II on a set of species • Run MP or ML on gene I and obtain a set U1 of trees, represented by its consensus tree t1 • Run MP or ML on gene II and obtain a set U2 of trees, represented by its consensus tree t2 • F ...
HealthGrid Conference
... Definition of the parameters for doing the Bayesian calculation Determination of the model of evolution Multiple alignment of the sequences previously to the final result Fig. 1 The Taverna workflow used for calculating with MrBayes ...
... Definition of the parameters for doing the Bayesian calculation Determination of the model of evolution Multiple alignment of the sequences previously to the final result Fig. 1 The Taverna workflow used for calculating with MrBayes ...
Key terms: Positional homology Homoplasy Reversal Parallelism
... understanding cases of evolutionary dissociation. Are there possible problems with using the concepts of serial homology, functional homology, and biological homology? 4. What is meant by the phrase “trees within trees”. How are the concepts of mutation, polymorphism and substitution related to this ...
... understanding cases of evolutionary dissociation. Are there possible problems with using the concepts of serial homology, functional homology, and biological homology? 4. What is meant by the phrase “trees within trees”. How are the concepts of mutation, polymorphism and substitution related to this ...
Classification of Bears
... (species, populations, individuals)--and branches, which define the relationship between the taxonomic units in terms of descent and ancestry. Only one branch can connect any two adjacent nodes. The branching pattern of the tree is called the topology and the branch length usually represents the num ...
... (species, populations, individuals)--and branches, which define the relationship between the taxonomic units in terms of descent and ancestry. Only one branch can connect any two adjacent nodes. The branching pattern of the tree is called the topology and the branch length usually represents the num ...
2_Outline_BIO119_div..
... B. Example: Genus, Species: Escherichia coli must be Latin endings. 1. Genus is always capitalized and the species is lower case 2. Always italicize or underline. 3. Name usually has some significance. C. How do identify a new isolate and classify it to the species level? 1. There are international ...
... B. Example: Genus, Species: Escherichia coli must be Latin endings. 1. Genus is always capitalized and the species is lower case 2. Always italicize or underline. 3. Name usually has some significance. C. How do identify a new isolate and classify it to the species level? 1. There are international ...
Exploratory Data Analysis Tools for Phylogenetics: Visualizing
... never have 3 mutually incompatible splits. Any split in the split system must be in at least 4 trees. ...
... never have 3 mutually incompatible splits. Any split in the split system must be in at least 4 trees. ...
Notes 3
... Phylogenetic systematics: The overall goal is to make higher categories reflect evolutionary history by requiring that all taxa be monophyletic. In a phylogenetic classification, species in the same genus had a most recent common ancestor more recently than species in different genera in the same fa ...
... Phylogenetic systematics: The overall goal is to make higher categories reflect evolutionary history by requiring that all taxa be monophyletic. In a phylogenetic classification, species in the same genus had a most recent common ancestor more recently than species in different genera in the same fa ...
phylogeny
... Phylogenetic systematics: The overall goal is to make higher categories reflect evolutionary history by requiring that all taxa be monophyletic. In a phylogenetic classification, species in the same genus had a most recent common ancestor more recently than species in different genera in the same fa ...
... Phylogenetic systematics: The overall goal is to make higher categories reflect evolutionary history by requiring that all taxa be monophyletic. In a phylogenetic classification, species in the same genus had a most recent common ancestor more recently than species in different genera in the same fa ...
Artificial Intelligence Project #3 : Analysis of Decision Tree Learning
... Each gene is probed by a set of short oligos. Each gene expression level is summarized by Signal: numerical value describing the abundance of mRNA A/P call: denotes the statistical significance of signal ...
... Each gene is probed by a set of short oligos. Each gene expression level is summarized by Signal: numerical value describing the abundance of mRNA A/P call: denotes the statistical significance of signal ...
Chapter 2: The Chemical Context of Life
... appearance (rapid environmental change leads to rapid evolution; also, small changes in genes can lead to large morphological differences) • Organisms that appear similar not always closely related (convergent evolution) • Just because 2 groups share primitive characters does not mean they are close ...
... appearance (rapid environmental change leads to rapid evolution; also, small changes in genes can lead to large morphological differences) • Organisms that appear similar not always closely related (convergent evolution) • Just because 2 groups share primitive characters does not mean they are close ...
such as for example in pairwise distance methods
... point are reconstructed • the minimum number of events to explain the sequence differences over the whole tree is computed: the minimum number of substitutions is computed for each nucleotide (or amino acid) site, and the numbers for all sites are added. • another tree topology is chosen ...
... point are reconstructed • the minimum number of events to explain the sequence differences over the whole tree is computed: the minimum number of substitutions is computed for each nucleotide (or amino acid) site, and the numbers for all sites are added. • another tree topology is chosen ...
macroevolutoin part i: phylogenies
... The following rules apply to reconstructing a phylogeny: 1. Maximum likelihood states that when considering multiple phylogenetic hypotheses, one should take into account the one that reflects the most likely sequence of evolutionary events given certain rules about how DNA changes over time. 2. Max ...
... The following rules apply to reconstructing a phylogeny: 1. Maximum likelihood states that when considering multiple phylogenetic hypotheses, one should take into account the one that reflects the most likely sequence of evolutionary events given certain rules about how DNA changes over time. 2. Max ...
Molecular Phylogeny
... • There are 3 main classes of phylogenetic methods for constructing phylogenies from sequence data : Methods directly based on sequences : • Maximum Parsimony : find a phylogenetic tree that explains the data, with as few evolutionary changes as possible. • Maximum likelihood : find a tree that maxi ...
... • There are 3 main classes of phylogenetic methods for constructing phylogenies from sequence data : Methods directly based on sequences : • Maximum Parsimony : find a phylogenetic tree that explains the data, with as few evolutionary changes as possible. • Maximum likelihood : find a tree that maxi ...
From phylogenetic trees to networks
... evolutionary history of genes, populations, species. They are typically reconstructed with a wide range of algorithms from the comparison of very long strings representing the molecular (DNA or protein) sequences found across different organisms. Phylogenetic reconstruction is part of the daily rout ...
... evolutionary history of genes, populations, species. They are typically reconstructed with a wide range of algorithms from the comparison of very long strings representing the molecular (DNA or protein) sequences found across different organisms. Phylogenetic reconstruction is part of the daily rout ...
Bioinformatics and Supercomputing
... •Discovery of Alu subfamillies led to hypothesis of master/ source genes. AGCT •Reveal ancestry because individuals only share particular sequence insertion if the share an ancestor. •Can identify similarities of functional, structural, or evolutionary relationships between the sequences ...
... •Discovery of Alu subfamillies led to hypothesis of master/ source genes. AGCT •Reveal ancestry because individuals only share particular sequence insertion if the share an ancestor. •Can identify similarities of functional, structural, or evolutionary relationships between the sequences ...
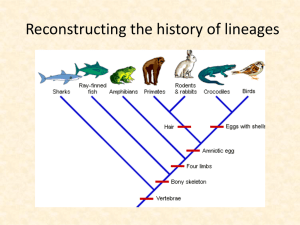
12-History of Lineages
... How can the history of evolution be reconstructed without seeing speciation events? 1) Identification of key characters (heritable parts or attributes of an organism) 2) Transform characters into transformation series 3) Compare these characters to an outgroup 4) Construct a cladogram based on the o ...
... How can the history of evolution be reconstructed without seeing speciation events? 1) Identification of key characters (heritable parts or attributes of an organism) 2) Transform characters into transformation series 3) Compare these characters to an outgroup 4) Construct a cladogram based on the o ...
Networks: expanding evolutionary thinking
... interpreting genomic data [10]. Tree-based genomic analysis is proving to be an accuracy challenge for the evolutionary biology community, and although genome-scale data carry the promise of fascinating insights into treelike processes, non-treelike processes are commonly observed. Network analysis ...
... interpreting genomic data [10]. Tree-based genomic analysis is proving to be an accuracy challenge for the evolutionary biology community, and although genome-scale data carry the promise of fascinating insights into treelike processes, non-treelike processes are commonly observed. Network analysis ...
Document
... Repeat this operation N times (100 or 1000 times if you can) Compute a consensus tree of the N trees Measure how many of the N trees agree with the consensus tree on each node ...
... Repeat this operation N times (100 or 1000 times if you can) Compute a consensus tree of the N trees Measure how many of the N trees agree with the consensus tree on each node ...