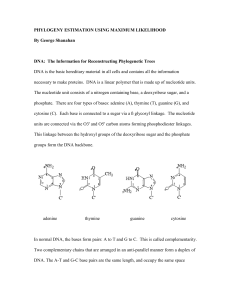
MultipleSequenceAlignment
... phylogenetic tree, aligns the most-alike pair, and incrementally adds sequences to the alignment in order of “alikeness” as indicated by the tree.) Differs from dynamic programming method for MSA in that it doesn’t refine the “first-cut” MSA by doing a full search through the reduced search space. ( ...
... phylogenetic tree, aligns the most-alike pair, and incrementally adds sequences to the alignment in order of “alikeness” as indicated by the tree.) Differs from dynamic programming method for MSA in that it doesn’t refine the “first-cut” MSA by doing a full search through the reduced search space. ( ...
Species tree
... • Bininda-Emonds ORP (2005). Supertree Construction in the Genomic Age. Methods in Enzymology 395: p.745-757. • Bininda-Emonds,OPRP, John L. Gittleman, Mike A. Steel (2002) The (super)Tree Of Life: Procedures, Problems, and Prospects. Annual Review of Ecology and Systematics, Vol. 33: 265-289. • Dag ...
... • Bininda-Emonds ORP (2005). Supertree Construction in the Genomic Age. Methods in Enzymology 395: p.745-757. • Bininda-Emonds,OPRP, John L. Gittleman, Mike A. Steel (2002) The (super)Tree Of Life: Procedures, Problems, and Prospects. Annual Review of Ecology and Systematics, Vol. 33: 265-289. • Dag ...
Use DNA Sequencing to Trace the Blue Whale`s Evolutionary Tree
... Although this is a genomics project, it is also about whales. To build the tree of the whale family, you will need to spend some time getting familiar with the scientific names and key features of about 25 whale and dolphin species. You will also learn about the blue whale's closest relatives that d ...
... Although this is a genomics project, it is also about whales. To build the tree of the whale family, you will need to spend some time getting familiar with the scientific names and key features of about 25 whale and dolphin species. You will also learn about the blue whale's closest relatives that d ...
C tudi - DNA to Darwin
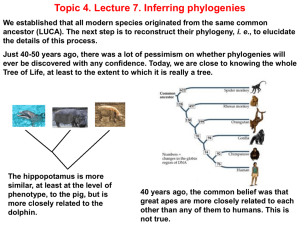
... a. Variations in the rate of evolution may lead to organisms being placed in the wrong place on an evolutionary tree (they may look very different when they are in fact closely-related). b. Any examples of convergent evolution could be suggested here, for example, wings in bats and birds, camera- ...
... a. Variations in the rate of evolution may lead to organisms being placed in the wrong place on an evolutionary tree (they may look very different when they are in fact closely-related). b. Any examples of convergent evolution could be suggested here, for example, wings in bats and birds, camera- ...
Matlab Bioinfo Toolbox QuickGuide
... to genomic and proteomic data formats, analysis techniques, and specialized visualizations for genomic and proteomic sequence and microarray analysis. The key features of the basic categories in the Bioinformatics Toolbox will be presented in the following sections. This guide does not replace the e ...
... to genomic and proteomic data formats, analysis techniques, and specialized visualizations for genomic and proteomic sequence and microarray analysis. The key features of the basic categories in the Bioinformatics Toolbox will be presented in the following sections. This guide does not replace the e ...
Chapter 25 Presentation
... For example, let’s look at hinged jaws. These are absent in lampreys, but are found in other members of the ingroup-this represents a branch point. The cladogram we’ve developed isn’t a phylogenetic tree, we need more information from fossils, etc. to indicate when the groups first appeared. ...
... For example, let’s look at hinged jaws. These are absent in lampreys, but are found in other members of the ingroup-this represents a branch point. The cladogram we’ve developed isn’t a phylogenetic tree, we need more information from fossils, etc. to indicate when the groups first appeared. ...
Phylogenetic Place of Guinea Pigs: No Support of the Rodent
... Graur et al’s ( 199 1) hypothesis that the guinea pig-like rodents have an evolutionary origin within mammals that is separate from that of other rodents (the rodent-polyphyly hypothesis) was reexamined by the maximum-likelihood method for protein phylogeny, as well as by the maximum-parsimony and n ...
... Graur et al’s ( 199 1) hypothesis that the guinea pig-like rodents have an evolutionary origin within mammals that is separate from that of other rodents (the rodent-polyphyly hypothesis) was reexamined by the maximum-likelihood method for protein phylogeny, as well as by the maximum-parsimony and n ...
1) of
... Often, data FAIL both tests - there are conflicts, and distances from sister species to the outgroup are not the same - revealing that evolution was not exclusively divergent and did not proceed at an exactly constant rate. What to do? More sophisticated approaches can be used. Maximal likelihood - ...
... Often, data FAIL both tests - there are conflicts, and distances from sister species to the outgroup are not the same - revealing that evolution was not exclusively divergent and did not proceed at an exactly constant rate. What to do? More sophisticated approaches can be used. Maximal likelihood - ...
Presentation Tuesday
... similarity. Fast, but not a direct reconstruction of “what happened in evolution”. Neighbor Joining is the often used method here ...
... similarity. Fast, but not a direct reconstruction of “what happened in evolution”. Neighbor Joining is the often used method here ...
Hemiplasy: A New Term in the Lexicon of Phylogenetics
... that can lead to genuine discordances between particular gene trees (components of the genome) and a composite or overall species phylogeny. We suggest the word hemiplasy, because the responsible lineage sorting processes have homoplasy-like consequences despite the fact that the character states th ...
... that can lead to genuine discordances between particular gene trees (components of the genome) and a composite or overall species phylogeny. We suggest the word hemiplasy, because the responsible lineage sorting processes have homoplasy-like consequences despite the fact that the character states th ...
Appendix S2.
... Most published trees contained only a few taxa of interest for any particular group. Therefore, we were often forced to use multiple trees which may have employed different characters and methods in their analyses to place particular taxa into our tree (e.g. Elapideae, see below). ...
... Most published trees contained only a few taxa of interest for any particular group. Therefore, we were often forced to use multiple trees which may have employed different characters and methods in their analyses to place particular taxa into our tree (e.g. Elapideae, see below). ...
A DNA-sequence based phylogeny for triculine snails (Gastropoda
... together with considerable bias among the 6 different types of nucleotide substitution (see Results). In such cases the ML method, making possible a fully optimized model of substitution, is considered more robust than other phylogenetic methods (Nei, 1991). A Bayesian method was also used because t ...
... together with considerable bias among the 6 different types of nucleotide substitution (see Results). In such cases the ML method, making possible a fully optimized model of substitution, is considered more robust than other phylogenetic methods (Nei, 1991). A Bayesian method was also used because t ...
TreeFitter commands
... Tree fitting has important applications in historical biogeography, coevolution and gene tree-species tree fitting [see recent reviews by \Page, 1998 #1419; Ronquist, 1998 #760]. A general characteristic of these problems is that two different kinds of trees are fitted to each other because we are i ...
... Tree fitting has important applications in historical biogeography, coevolution and gene tree-species tree fitting [see recent reviews by \Page, 1998 #1419; Ronquist, 1998 #760]. A general characteristic of these problems is that two different kinds of trees are fitted to each other because we are i ...
Molecular phylogeny, part B
... Multigene family: A group of genes, clustered or dispersed, with related nucleotide sequences. Multiple alignment: An alignment of three or more nucleotide sequences. Multiple hit or multiple substitution: The situation that occurs when a single nucleotide in a DNA sequence undergoes two mutational ...
... Multigene family: A group of genes, clustered or dispersed, with related nucleotide sequences. Multiple alignment: An alignment of three or more nucleotide sequences. Multiple hit or multiple substitution: The situation that occurs when a single nucleotide in a DNA sequence undergoes two mutational ...
Classification and Phylogeny
... The ancestral state of character j is present in species 2. This is an evolutionary reversal. ...
... The ancestral state of character j is present in species 2. This is an evolutionary reversal. ...
PowerPoint
... 2. Parsimony – fewest number of evolutionary events (mutations) – relatively often fails to reconstruct correct phylogeny, but methods have improved recently 3. Maximum likelihood – L = Pr[Data|Tree] – most flexible class of methods - user-specified evolutionary methods can be used ...
... 2. Parsimony – fewest number of evolutionary events (mutations) – relatively often fails to reconstruct correct phylogeny, but methods have improved recently 3. Maximum likelihood – L = Pr[Data|Tree] – most flexible class of methods - user-specified evolutionary methods can be used ...
Concepts of Biology - Amazon Simple Storage Service (S3)
... other organisms not pictured, such as fungi and protists. At each sublevel, the organisms become more similar because they are more closely related. Before Darwin’s theory of evolution was developed, naturalists sometimes classified organisms using arbitrary similarities, but since the theory of evo ...
... other organisms not pictured, such as fungi and protists. At each sublevel, the organisms become more similar because they are more closely related. Before Darwin’s theory of evolution was developed, naturalists sometimes classified organisms using arbitrary similarities, but since the theory of evo ...
Amsterdam 2004 - Theoretical Biology & Bioinformatics
... multidomain proteins by examining the pictorial representation of the BLAST search outputs. The sequences of detected multidomain proteins are split into single-domain segments and steps 1–4 are repeated with these sequences, which results in the assignment of individual domains to COGs in accordanc ...
... multidomain proteins by examining the pictorial representation of the BLAST search outputs. The sequences of detected multidomain proteins are split into single-domain segments and steps 1–4 are repeated with these sequences, which results in the assignment of individual domains to COGs in accordanc ...
The Graph of Life
... •The three trees seem quite different: (((((((Scer,Spar),Smik),Skud),Sbay),Scas),Sklu),Calb) (((((((Scer,Spar),Smik),Skud),Sbay),Sklu),Scas),Calb) (((((Skud,Sbay),((Scer,Spar),Smik)),Scas),Sklu),Calb) In particular, Skud seems to move a lot. But our graph showed multiple ancestry for Scas only. ...
... •The three trees seem quite different: (((((((Scer,Spar),Smik),Skud),Sbay),Scas),Sklu),Calb) (((((((Scer,Spar),Smik),Skud),Sbay),Sklu),Scas),Calb) (((((Skud,Sbay),((Scer,Spar),Smik)),Scas),Sklu),Calb) In particular, Skud seems to move a lot. But our graph showed multiple ancestry for Scas only. ...
Unoshan_project
... but they are not in the usual Watson-Crick geometry. Sequences can diverge from a common ancestor because mutations occur. Those mutations can then be fixed into the evolving population. The Maximum Likelihood method is used for the analysis of DNA and amino acid sequence data in an attempt to answe ...
... but they are not in the usual Watson-Crick geometry. Sequences can diverge from a common ancestor because mutations occur. Those mutations can then be fixed into the evolving population. The Maximum Likelihood method is used for the analysis of DNA and amino acid sequence data in an attempt to answe ...
25_DetailLectOutjk_AR
... In general, the more points of resemblance that two complex structures have, the less likely it is that they evolved independently. For example, the skulls of a human and a chimpanzee are formed by the fusion of many bones. The two skulls match almost perfectly, bone for bone. It is highly unl ...
... In general, the more points of resemblance that two complex structures have, the less likely it is that they evolved independently. For example, the skulls of a human and a chimpanzee are formed by the fusion of many bones. The two skulls match almost perfectly, bone for bone. It is highly unl ...
PowerPoint
... Consider the codons specifying aspartic acid and lysine: both start AA, lysine ends A or G, and aspartic acid ends T or C. So, if the rate at which C changes to T is higher from the rate that C changes to G or A (as is often the case), then more of the changes at the third position will be synonymou ...
... Consider the codons specifying aspartic acid and lysine: both start AA, lysine ends A or G, and aspartic acid ends T or C. So, if the rate at which C changes to T is higher from the rate that C changes to G or A (as is often the case), then more of the changes at the third position will be synonymou ...
RidgeRace: ridge regression for continuous ancestral character
... and Contml (Felsenstein, 1993). One of the simplest ways to reconstruct a continuous ancestral character state was established with Felsenstein’s algorithm for ‘Phylogenetic Independent Contrasts’ (Felsenstein, 1985). In the Phylogenetic Independent Contrasts algorithm, ancestral values are computed ...
... and Contml (Felsenstein, 1993). One of the simplest ways to reconstruct a continuous ancestral character state was established with Felsenstein’s algorithm for ‘Phylogenetic Independent Contrasts’ (Felsenstein, 1985). In the Phylogenetic Independent Contrasts algorithm, ancestral values are computed ...