Phylogenetic Analysis
... (2) Calculate distance from x and y to their ancestor z by 3-leaf tree formula (3) Remove the rows and columns of x and y in D (4) Insert the row and column of z with distance by 3-leaf tree formula (5) Repeat (1)~(4) until completes an unrooted tree ...
... (2) Calculate distance from x and y to their ancestor z by 3-leaf tree formula (3) Remove the rows and columns of x and y in D (4) Insert the row and column of z with distance by 3-leaf tree formula (5) Repeat (1)~(4) until completes an unrooted tree ...
Evolution/Phylogeny
... Note that with evolutionary methods for generating trees you get distances between objects by walking from one to the other. ...
... Note that with evolutionary methods for generating trees you get distances between objects by walking from one to the other. ...
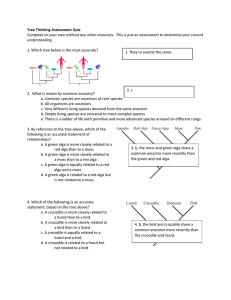
Tree Thinking Assessment Quiz
... frog groups with branches drawn proportional to absolute time. Error bars on internal nodes depict confidence intervals on the dates of estimated nodes. Assuming this tree and the associated ages are correct which of the following statements is true? a. No individual living before 70 million 16. d y ...
... frog groups with branches drawn proportional to absolute time. Error bars on internal nodes depict confidence intervals on the dates of estimated nodes. Assuming this tree and the associated ages are correct which of the following statements is true? a. No individual living before 70 million 16. d y ...
Practice exam questions
... which you will have a selection of possible answers provided and you pick the correct one (as in quizler), and some in which you match a term to a definition or a scientist to a given scientific discovery or evolutionary insight. You will have a selection of questions on the last page, from which yo ...
... which you will have a selection of possible answers provided and you pick the correct one (as in quizler), and some in which you match a term to a definition or a scientist to a given scientific discovery or evolutionary insight. You will have a selection of questions on the last page, from which yo ...
here - CMBI
... sequence changes to other character states • BLOck SUbstitution Matrix (BLOSUM) is based on observed substitutions between proteins with e.g. >62% sequence identity ...
... sequence changes to other character states • BLOck SUbstitution Matrix (BLOSUM) is based on observed substitutions between proteins with e.g. >62% sequence identity ...
Practical theory (15-20 min) A phylogeny is the representation of the
... “Phylogenetic Tree” allows the user to visualize and download the built tree. It could be visualized as a cladogram (“Cladogram”) or as a Phylogram (“Real”), depending on the user's needs. When a branch has length = 0, those sequences are exactly the same. The bigger the distance is, the more separa ...
... “Phylogenetic Tree” allows the user to visualize and download the built tree. It could be visualized as a cladogram (“Cladogram”) or as a Phylogram (“Real”), depending on the user's needs. When a branch has length = 0, those sequences are exactly the same. The bigger the distance is, the more separa ...
Molecular Phylogenetic Analysis: Design and Implementation of
... thousand sequences), the update processes should happen at most every three or four months. Meanwhile, the new sequences that might provide relevant information for biological studies are hold until the next reconstruction. We proposed PHYSER [10] as a new method to detect possible sequencing errors ...
... thousand sequences), the update processes should happen at most every three or four months. Meanwhile, the new sequences that might provide relevant information for biological studies are hold until the next reconstruction. We proposed PHYSER [10] as a new method to detect possible sequencing errors ...
Phylogeography
... searching tree space Local optima are problem: need to traverse valleys to get to other peaks Heuristic search: cut trees up systematically and reassemble Branch and bound: search for optimal path through tree space ...
... searching tree space Local optima are problem: need to traverse valleys to get to other peaks Heuristic search: cut trees up systematically and reassemble Branch and bound: search for optimal path through tree space ...
A method for paralogy trees reconstruction
... Genes belonging to the same organism are called paralogs when they show a significant similarity in the sequences, even if they have a different biological function. It is an emergent biological paradigm that the families of paralogs derive from a mechanism of gene duplication with modification, rep ...
... Genes belonging to the same organism are called paralogs when they show a significant similarity in the sequences, even if they have a different biological function. It is an emergent biological paradigm that the families of paralogs derive from a mechanism of gene duplication with modification, rep ...
S6. Phylogenetic results: complementary analyses Bayesian
... keeping the (relatively short) 16S rRNA fragment as one partition and defining each codon position of ND2 as separate partition (S6b). For the nDNA dataset, we used a scheme with 3 partitions (each codon position of the combined nuclear genes defined as separate partition; S6c) and one with 12 parti ...
... keeping the (relatively short) 16S rRNA fragment as one partition and defining each codon position of ND2 as separate partition (S6b). For the nDNA dataset, we used a scheme with 3 partitions (each codon position of the combined nuclear genes defined as separate partition; S6c) and one with 12 parti ...
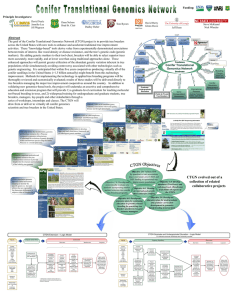
Objective 2.0
... across the United States with new tools to enhance and accelerate traditional tree improvement activities. These “knowledge-based” tools derive value from experimentally demonstrated associations between traits of interest, like wood density or disease resistance, and the tree’s genetic code (geneti ...
... across the United States with new tools to enhance and accelerate traditional tree improvement activities. These “knowledge-based” tools derive value from experimentally demonstrated associations between traits of interest, like wood density or disease resistance, and the tree’s genetic code (geneti ...
PPT - UT Computer Science
... • Much software exists, most of which attempt to solve one of two major optimization criteria: Maximum Parsimony and Maximum Likelihood. The most frequently used software package is PAUP*, which contains many different heuristics. • Methods for phylogeny reconstruction are evaluated primarily in sim ...
... • Much software exists, most of which attempt to solve one of two major optimization criteria: Maximum Parsimony and Maximum Likelihood. The most frequently used software package is PAUP*, which contains many different heuristics. • Methods for phylogeny reconstruction are evaluated primarily in sim ...
Phylogenetics
... Phylogenetics exercise, Bioinformatics for cell biologists, 2011 This exercise will simulate the construction of a phylogenetic tree for organisms where only individual genes (rather than whole genomes) are sequenced, akin to the construction of 16S rRNA-based trees for bacteria. The same tools can ...
... Phylogenetics exercise, Bioinformatics for cell biologists, 2011 This exercise will simulate the construction of a phylogenetic tree for organisms where only individual genes (rather than whole genomes) are sequenced, akin to the construction of 16S rRNA-based trees for bacteria. The same tools can ...
BIOLOGY - Learner
... Although the methods used in cladistic analysis are the same for both molecular and morphological characters, molecular data provides several advantages. First, molecular data offers a large and essentially limitless set of characters. Each nucleotide position, in theory, can be considered a charact ...
... Although the methods used in cladistic analysis are the same for both molecular and morphological characters, molecular data provides several advantages. First, molecular data offers a large and essentially limitless set of characters. Each nucleotide position, in theory, can be considered a charact ...
No Slide Title
... cause problems for phylogenetic analysis • Felsenstein (1978) made a simple model phylogeny including four taxa and a mixture of short and long branches A p TRUE TREE ...
... cause problems for phylogenetic analysis • Felsenstein (1978) made a simple model phylogeny including four taxa and a mixture of short and long branches A p TRUE TREE ...
Many of the slides that I`ll use have been borrowed from Dr. Paul
... If the rules are valid (logically sound) and the premises are true, then the conclusions are guaranteed to be true. ...
... If the rules are valid (logically sound) and the premises are true, then the conclusions are guaranteed to be true. ...
Maximum pseudo-likelihood estimation of species trees (MP
... NTAXA to 300. MAXROUND defines the maximum number of rounds the algorithm will run. The algorithm will be terminated when the MAXROUNDth round is reached. If you think the current setting MAXROUND = 10000000 is too small for the algorithm to find the maximum pseudo-likelihood estimate of the species ...
... NTAXA to 300. MAXROUND defines the maximum number of rounds the algorithm will run. The algorithm will be terminated when the MAXROUNDth round is reached. If you think the current setting MAXROUND = 10000000 is too small for the algorithm to find the maximum pseudo-likelihood estimate of the species ...
Systematics and phylogeny
... Early trees • Similarity may not accurately predict evolutionary relationships – Early systematists relied on the expectation that the greater the time since two species diverged from a common ancestor, more different would be Evolution can happen quickly ...
... Early trees • Similarity may not accurately predict evolutionary relationships – Early systematists relied on the expectation that the greater the time since two species diverged from a common ancestor, more different would be Evolution can happen quickly ...
BIOL2007 - EVOLUTIONARY TREES AND THEIR USES
... 2) Most use ‘Tree-searching’ methods – compare large sample of trees from among huge number possible for large data-sets. Use computerised search routines to maximise chance of finding shortest trees compatible with the data. Aim is to assess of reliability of shortest tree compared to other trees ...
... 2) Most use ‘Tree-searching’ methods – compare large sample of trees from among huge number possible for large data-sets. Use computerised search routines to maximise chance of finding shortest trees compatible with the data. Aim is to assess of reliability of shortest tree compared to other trees ...
Phylogeny
... • Clustalw: very simple, not very sophisticated (best avoided for phylogeny, s.above) easily obtained/learned • Phylip: very powerful, less convivial • TreeTop (Genebee server): is a web server that produces high quality trees ...
... • Clustalw: very simple, not very sophisticated (best avoided for phylogeny, s.above) easily obtained/learned • Phylip: very powerful, less convivial • TreeTop (Genebee server): is a web server that produces high quality trees ...
biol2007 - evolutionary trees and their uses
... species. Which tree needs fewest evolutionary changes? Choose a tree. Plot onto it, using parsimony, positions at which each character must have changed. e.g. Tree 1, character a: state in Species 1 & 2 & 3 = A, state in 4 & 5 = C. We can infer there was a change from C to A in the common ancestor o ...
... species. Which tree needs fewest evolutionary changes? Choose a tree. Plot onto it, using parsimony, positions at which each character must have changed. e.g. Tree 1, character a: state in Species 1 & 2 & 3 = A, state in 4 & 5 = C. We can infer there was a change from C to A in the common ancestor o ...
phylogenetic tree.
... Can you mark the point where all carnivores shared a common ancestor? Trees show patterns of descent, not phenotypic similarity. They do not show the age of the particular species. ...
... Can you mark the point where all carnivores shared a common ancestor? Trees show patterns of descent, not phenotypic similarity. They do not show the age of the particular species. ...
PHYLOGENY AND EVOLUTION OF CORNALES: INTEGRATING
... Recent advances in plant phylogenetic studies revealed that the dogwoods and close relatives (order Cornales) represent an ancient evolutionary lineage within the most diverse and highly specialized group of flowering plants, the asterid group. The phylogenetic position of Cornales makes the order c ...
... Recent advances in plant phylogenetic studies revealed that the dogwoods and close relatives (order Cornales) represent an ancient evolutionary lineage within the most diverse and highly specialized group of flowering plants, the asterid group. The phylogenetic position of Cornales makes the order c ...
Document
... characters? • How do we sort out phylogeny from a mixture of signal (synapomorphies) and noise (homoplasy). • Cladistic methodology (Willi Hennig) utilizes the principle of ...
... characters? • How do we sort out phylogeny from a mixture of signal (synapomorphies) and noise (homoplasy). • Cladistic methodology (Willi Hennig) utilizes the principle of ...