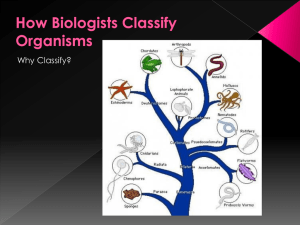
Phylogenies show Evolutionary Relationships
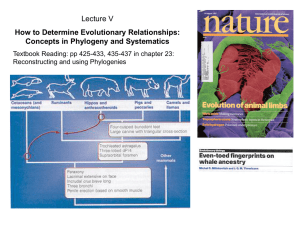
... shared ancestry, such as the bones of a whale’s flipper and a tiger’s paw -They are used to construct phylogenetic trees, the more structures you share with another organism, the closer you are on the tree because its more likely you share a common ancestor -As new evidence about organisms becomes a ...
... shared ancestry, such as the bones of a whale’s flipper and a tiger’s paw -They are used to construct phylogenetic trees, the more structures you share with another organism, the closer you are on the tree because its more likely you share a common ancestor -As new evidence about organisms becomes a ...
C.Constance Biol 415 Hiram College
... 1904: Nuttall used immunological tests to deduce relationships between a variety of animals to place humans in their correct evolutionary position relative to other primates ...
... 1904: Nuttall used immunological tests to deduce relationships between a variety of animals to place humans in their correct evolutionary position relative to other primates ...
File - Down the Rabbit Hole
... nested groups, in which similar or related groups at one level are combined into larger and more general groups at the next higher level. Biological classification is based on shared descent from the nearest common ancestor ...
... nested groups, in which similar or related groups at one level are combined into larger and more general groups at the next higher level. Biological classification is based on shared descent from the nearest common ancestor ...
Lecture PPT - Carol Lee Lab
... • Synapomorphies are shared derived homologous traits • They can be DNA nucleotides or other heritable traits • They are used to group taxa that are more closely related to one another ...
... • Synapomorphies are shared derived homologous traits • They can be DNA nucleotides or other heritable traits • They are used to group taxa that are more closely related to one another ...
Lecture PPT - Carol Lee Lab
... • Synapomorphies are shared derived homologous traits • They can be DNA nucleotides or other heritable traits • They are used to group taxa that are more closely related to one another ...
... • Synapomorphies are shared derived homologous traits • They can be DNA nucleotides or other heritable traits • They are used to group taxa that are more closely related to one another ...
File - Ms. Poole`s Biology
... classification of carnivores, however with the advancements in DNA and protein analysis, changes have been made in the traditional classification of organisms and their phylogeny. ...
... classification of carnivores, however with the advancements in DNA and protein analysis, changes have been made in the traditional classification of organisms and their phylogeny. ...
Chapter 25
... species and all of its descendents paraphyletic – made up of an ancestral species and only some of its descendents polyphyletic – a grouping that leaves out the common ancestor of the species included ...
... species and all of its descendents paraphyletic – made up of an ancestral species and only some of its descendents polyphyletic – a grouping that leaves out the common ancestor of the species included ...
Phylogeny slides
... Brute force algorithm: consider all possible alignments, then determine the one that results in a best score (time complexity?) Common Heuristic: Use regular (two-string) alignment and then repeatedly add a string to a growing alignment ...
... Brute force algorithm: consider all possible alignments, then determine the one that results in a best score (time complexity?) Common Heuristic: Use regular (two-string) alignment and then repeatedly add a string to a growing alignment ...
tree - Tecfa
... Character-based (Sequence) methods 1. Constructs a phylogenetic tree based on the ...
... Character-based (Sequence) methods 1. Constructs a phylogenetic tree based on the ...
No Slide Title - Faculty Virginia
... Systematists classify organisms and determine evolutionary relationships based on analysis of homologous characters (traits) •Systematic investigation is based on analysis of homologous characters (traits); characters may be morphological, molecular, behavioral, physiological.. •Homologous charac ...
... Systematists classify organisms and determine evolutionary relationships based on analysis of homologous characters (traits) •Systematic investigation is based on analysis of homologous characters (traits); characters may be morphological, molecular, behavioral, physiological.. •Homologous charac ...
Two-point Linkage Analysis: a brief outline of theory
... Topology: the branching patterns of the tree Branch length (scaled trees only): represents the number of changes that have occurred in the branch Root: the common ancestor of all taxa Clade: a group of two or more taxa or DNA sequences that ...
... Topology: the branching patterns of the tree Branch length (scaled trees only): represents the number of changes that have occurred in the branch Root: the common ancestor of all taxa Clade: a group of two or more taxa or DNA sequences that ...
Chapter 25
... organisms into increasingly broad taxonomic categories based on their similarities and differences with respect to a set of characteristics. Systematics is an analytical approach to understanding the diversity and relationships of living and extinct organisms based on similarities and differences an ...
... organisms into increasingly broad taxonomic categories based on their similarities and differences with respect to a set of characteristics. Systematics is an analytical approach to understanding the diversity and relationships of living and extinct organisms based on similarities and differences an ...
Intraspecific gene genealogies: trees grafting into networks
... low divergence -> fewer characters for phylogenetic analysis ...
... low divergence -> fewer characters for phylogenetic analysis ...
Genealogical Trees,Coalescent Theory and the Analysis of Genetic
... Since Polymorphisms are Random Processes, then they can be studied by their statistical properties ...
... Since Polymorphisms are Random Processes, then they can be studied by their statistical properties ...
Evolution of HSV-1 and VZV.
... -A way to get “statistical significance” of a certain topology -Construct several new sequence sets (1000 st.) ...
... -A way to get “statistical significance” of a certain topology -Construct several new sequence sets (1000 st.) ...
View the seminar poster
... late 1990s, early studies drew on small sets of external morphological characters, mostly those used in classical taxonomic works. In order to bolster the character sample, new anatomical data were worked ...
... late 1990s, early studies drew on small sets of external morphological characters, mostly those used in classical taxonomic works. In order to bolster the character sample, new anatomical data were worked ...
BlueJam Evolutionary Music Composition
... all kinds. Their approach to problem solving can be described as a search through the space of possible solutions. In 1992, Koza brought Tree-based GP to prominence, and it has been used widely ever since. The same paradigm is followed with Heuristic Trees, which uses the structure of the tree to sp ...
... all kinds. Their approach to problem solving can be described as a search through the space of possible solutions. In 1992, Koza brought Tree-based GP to prominence, and it has been used widely ever since. The same paradigm is followed with Heuristic Trees, which uses the structure of the tree to sp ...
Comparative Anatomy: Phylogenetics Assignment
... two hypotheses (one from Neighbor-joining algorithm and the other from maximum parsimony) derived from sequence data from the 28s rRNA gene of the mitochondrial genome, both did not resolve satisfactory relationships among our fishes for various reasons. The goal of this section is for you to try on ...
... two hypotheses (one from Neighbor-joining algorithm and the other from maximum parsimony) derived from sequence data from the 28s rRNA gene of the mitochondrial genome, both did not resolve satisfactory relationships among our fishes for various reasons. The goal of this section is for you to try on ...
Slides of Barbara`s talk - School of Mathematical Sciences
... The MP criterion has been shown to be statistically inconsistent on some trees under the models of nucleotide substitution discussed previously. Likelihood is statisitically consistent (given the correct model). ...
... The MP criterion has been shown to be statistically inconsistent on some trees under the models of nucleotide substitution discussed previously. Likelihood is statisitically consistent (given the correct model). ...
Phylogenetic analysis of flatfish species (Teleostei
... one species P. pinnifasciatus. Other species, placed in different genera Pseudopleuronectes, Liopsetta, and Lepidopsetta. The diagnostics of species (barcoding) based on Co-1 and Cyt- b gene sequences is highly effective because of low intraspecies and high interspecies variability of this markers ...
... one species P. pinnifasciatus. Other species, placed in different genera Pseudopleuronectes, Liopsetta, and Lepidopsetta. The diagnostics of species (barcoding) based on Co-1 and Cyt- b gene sequences is highly effective because of low intraspecies and high interspecies variability of this markers ...
Our material on phylogenetics in bioinformatics was roughly divided
... * what is the likelihood ratio test and how is it used to test a wide variety of possible hypotheses about sequence evolution, such as: rates of evolution, monophyly of group or sequences, similarity of branching history of two trees, etc. ...
... * what is the likelihood ratio test and how is it used to test a wide variety of possible hypotheses about sequence evolution, such as: rates of evolution, monophyly of group or sequences, similarity of branching history of two trees, etc. ...
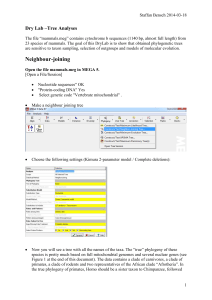
Dry Lab – More Tree Analayses
... One of the traditional and still very popular discrete method is Maximum Parsimony. Go to “Phylogeny/ Test Maximum Parsimony Trees”. Use the default settings [(CNI (level=1) with initial tree by Random addition (10 reps))] and select bootstrapping. Compare how much longer it takes to do bootstraps w ...
... One of the traditional and still very popular discrete method is Maximum Parsimony. Go to “Phylogeny/ Test Maximum Parsimony Trees”. Use the default settings [(CNI (level=1) with initial tree by Random addition (10 reps))] and select bootstrapping. Compare how much longer it takes to do bootstraps w ...
SK_DifficultProblems.
... Saturation – the problem of multiple changes at the same sites • Theory, simulations, and practical experience all indicate that the sequences must eventually lose information about events that were long ago. • Part of the problem with using DNA sequence alignments to infer deep events is that the ...
... Saturation – the problem of multiple changes at the same sites • Theory, simulations, and practical experience all indicate that the sequences must eventually lose information about events that were long ago. • Part of the problem with using DNA sequence alignments to infer deep events is that the ...