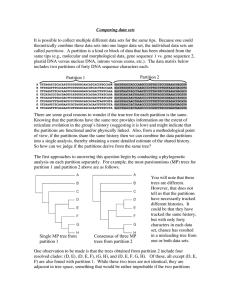
Comparing data sets It is possible to collect multiple different data
... particular clades. Further, recall that we can use a topology test to ask whether a tree with (or Optimal tree without) a particular clade can be rejected by a for part. 1 data set. If partition one rejects all trees than lack Plausible trees for part. 1 clade x, while partition two rejects all tree ...
... particular clades. Further, recall that we can use a topology test to ask whether a tree with (or Optimal tree without) a particular clade can be rejected by a for part. 1 data set. If partition one rejects all trees than lack Plausible trees for part. 1 clade x, while partition two rejects all tree ...
Supplementary Methods
... NGS experiments may produce vastly different numbers of total reads for paired data, e.g. YY1 ChIP-seq vs. Input. Suppose there are M reads for IP and N reads for Input. Then, many researchers currently scale the Input counts per genomic window by M/N, in order to equalize the total number of reads ...
... NGS experiments may produce vastly different numbers of total reads for paired data, e.g. YY1 ChIP-seq vs. Input. Suppose there are M reads for IP and N reads for Input. Then, many researchers currently scale the Input counts per genomic window by M/N, in order to equalize the total number of reads ...
Fieldwork in cultural Anthropology: Methods and Ethics
... LEARNING OBJECTIVES 1. Identify major ethnographic techniques & what type of information is gained from each technique. 2. Describe ethnographic techniques by identifying specific anthropological examples. 3. Summarize the methods used by anthropologists to study human populations. 4. Identify the ...
... LEARNING OBJECTIVES 1. Identify major ethnographic techniques & what type of information is gained from each technique. 2. Describe ethnographic techniques by identifying specific anthropological examples. 3. Summarize the methods used by anthropologists to study human populations. 4. Identify the ...
Appendix S3: Coalescent divergence time estimation: IMa2
... to estimate the posterior density of the parameters [1-3]. IMA2 also estimates the TMRCA from the many genealogies sampled. We ran IMA2 with simple two-population models, although the program accommodates >1 ancestral population thus >2 modern populations, because we analyzed a single locus, and the ...
... to estimate the posterior density of the parameters [1-3]. IMA2 also estimates the TMRCA from the many genealogies sampled. We ran IMA2 with simple two-population models, although the program accommodates >1 ancestral population thus >2 modern populations, because we analyzed a single locus, and the ...
You can position your opening statement here, either in
... One of the most widely used and practical forms of machine learning and data mining ...
... One of the most widely used and practical forms of machine learning and data mining ...
BIOL2007 - EVOLUTIONARY TREES AND THEIR USES
... that direction of change was from A to C because requires 2 changes in evolution of species 4 and 5 – not parsimonious Repeat for all characters to complete tree 1. Repeat process (usually via computer) for every possible tree. Somewhat tedious. Finally, count number of state changes (“length”) of e ...
... that direction of change was from A to C because requires 2 changes in evolution of species 4 and 5 – not parsimonious Repeat for all characters to complete tree 1. Repeat process (usually via computer) for every possible tree. Somewhat tedious. Finally, count number of state changes (“length”) of e ...
PPTX - Tandy Warnow
... How well do POY and BeeTLe do, compared to other MSA methods? • We simulated sequences down evolutionary trees with substitutions, insertions, and indels. • We computed alignments on each dataset using multiple techniques (e.g., POY, BeeTLe, Muscle, ...
... How well do POY and BeeTLe do, compared to other MSA methods? • We simulated sequences down evolutionary trees with substitutions, insertions, and indels. • We computed alignments on each dataset using multiple techniques (e.g., POY, BeeTLe, Muscle, ...
Phylogenetic Place of Guinea Pigs: No Support of the Rodent
... Graur et al’s ( 199 1) hypothesis that the guinea pig-like rodents have an evolutionary origin within mammals that is separate from that of other rodents (the rodent-polyphyly hypothesis) was reexamined by the maximum-likelihood method for protein phylogeny, as well as by the maximum-parsimony and n ...
... Graur et al’s ( 199 1) hypothesis that the guinea pig-like rodents have an evolutionary origin within mammals that is separate from that of other rodents (the rodent-polyphyly hypothesis) was reexamined by the maximum-likelihood method for protein phylogeny, as well as by the maximum-parsimony and n ...
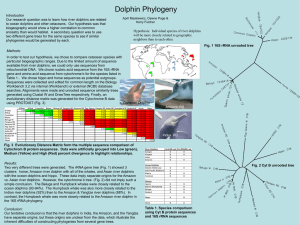
dolphin1
... Our research question was to learn how river dolphins are related to ocean dolphins and other cetaceans. Our hypothesis was that biogeography would show a higher correlation to common ancestry than would habitat. A secondary question was to use two different gene trees for the same species to see if ...
... Our research question was to learn how river dolphins are related to ocean dolphins and other cetaceans. Our hypothesis was that biogeography would show a higher correlation to common ancestry than would habitat. A secondary question was to use two different gene trees for the same species to see if ...
Comparison of different non-statistical classification
... The algorithm is extensively described in [20]. Further, it is extended to generate numeric constants by using two-level grammatical evolution as described in [21]. This is important when solving this classification problem, because the conditions are to be in format “if (age > 20) then will-sign el ...
... The algorithm is extensively described in [20]. Further, it is extended to generate numeric constants by using two-level grammatical evolution as described in [21]. This is important when solving this classification problem, because the conditions are to be in format “if (age > 20) then will-sign el ...
生物計算
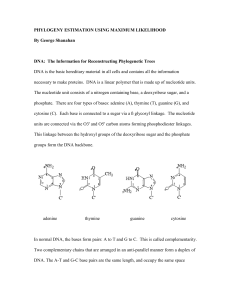
... at least two different nucleotides each of these nucleotides to present at least twice. ...
... at least two different nucleotides each of these nucleotides to present at least twice. ...
Unoshan_project
... Molecular phylogeny methods allow, from a given set of aligned sequences, the suggestion of phylogenetic trees which aim at reconstructing the history of successive divergence which took place during evolution, between the considered sequences and their common ancestor. Its topology (form) and its l ...
... Molecular phylogeny methods allow, from a given set of aligned sequences, the suggestion of phylogenetic trees which aim at reconstructing the history of successive divergence which took place during evolution, between the considered sequences and their common ancestor. Its topology (form) and its l ...
Document
... Problem: Long Branch Attraction (LBA) • Particular problem associated with parsimony methods • Rapidly evolving taxa are placed together in a tree regardless of their true position • Partly due to assumption in parsimony that all lineages evolve at the same rate • This means that also UPGMA suffers ...
... Problem: Long Branch Attraction (LBA) • Particular problem associated with parsimony methods • Rapidly evolving taxa are placed together in a tree regardless of their true position • Partly due to assumption in parsimony that all lineages evolve at the same rate • This means that also UPGMA suffers ...
Metabolomics meets Genomics
... errors such as self-reported relationships that are not accurate. The relationship errors can be corrected by consulting with the self-reported relationships and/or using inferred genetic relationships. ...
... errors such as self-reported relationships that are not accurate. The relationship errors can be corrected by consulting with the self-reported relationships and/or using inferred genetic relationships. ...
Studying the evolution of photosynthesis using phylogenetic trees
... is vivid as to which methods represent the underlying evolutionary families and relationships best [6, 7]. These tree maps can however be used as well to map the evolution of subsystems of living beings, the example of photosynthesis being discussed in the following [5]. The authors constructed phyl ...
... is vivid as to which methods represent the underlying evolutionary families and relationships best [6, 7]. These tree maps can however be used as well to map the evolution of subsystems of living beings, the example of photosynthesis being discussed in the following [5]. The authors constructed phyl ...
Systematics and phylogeny
... – Bacteria-----whales----sequoia trees • Biologists group organisms based on shared characteristics Taxonomy • Field of biology concerned with identifying and naming • Binomial system devised by Linnaeus • Classification is how species and higher groups are placed into the taxonomic hierarchy System ...
... – Bacteria-----whales----sequoia trees • Biologists group organisms based on shared characteristics Taxonomy • Field of biology concerned with identifying and naming • Binomial system devised by Linnaeus • Classification is how species and higher groups are placed into the taxonomic hierarchy System ...
view
... While the NN analysis detected significant effects, SVM analysis did have higher predictive accuracy. This might be explained by the limited number of architecture evaluated in the NN analyses. Nearly every paper discussed in this study claim that NN appear to be a good approach for gene mapping stu ...
... While the NN analysis detected significant effects, SVM analysis did have higher predictive accuracy. This might be explained by the limited number of architecture evaluated in the NN analyses. Nearly every paper discussed in this study claim that NN appear to be a good approach for gene mapping stu ...
Practice exam questions
... A. From the following terms, select the correct one to complete the sentences below. There are more terms than sentences so not all are used, and some terms might be used more than once. analogous apomorphic arthropods bilateral chordates cladogram DNA echinoderms guinea pigs ingroup mitochondrial g ...
... A. From the following terms, select the correct one to complete the sentences below. There are more terms than sentences so not all are used, and some terms might be used more than once. analogous apomorphic arthropods bilateral chordates cladogram DNA echinoderms guinea pigs ingroup mitochondrial g ...
Phylogeography
... Many algorithms exist for searching tree space Local optima are problem: need to traverse valleys to get to other peaks Heuristic search: cut trees up systematically and reassemble Branch and bound: search for optimal path through tree space ...
... Many algorithms exist for searching tree space Local optima are problem: need to traverse valleys to get to other peaks Heuristic search: cut trees up systematically and reassemble Branch and bound: search for optimal path through tree space ...
Lecture 4: (Part 1) Phylogenetic inference
... 1. Tree should reflect overall degree of similarity. 2. Tree should be based on as many characters as possible. 3. Tree should minimize the distance among taxa. ...
... 1. Tree should reflect overall degree of similarity. 2. Tree should be based on as many characters as possible. 3. Tree should minimize the distance among taxa. ...
In the article entitled ‘Search for a Tree of Life... evolution, at least as far as bacteria and archaea are
... (NUTs; that is, the trees for genes that are represented in all or nearly all archaea and bacteria), which include primarily genes for key protein components of the trans lation and transcription systems, showed particularly high topological similarity to the other trees in the FOL. Although the to ...
... (NUTs; that is, the trees for genes that are represented in all or nearly all archaea and bacteria), which include primarily genes for key protein components of the trans lation and transcription systems, showed particularly high topological similarity to the other trees in the FOL. Although the to ...
Branching Activity Instructions:
... What would it mean if a species’ line stopped before it got to the top? Which two species on the branching tree are the most closely related (have the most in common with each other)? Which two species have the least in common? ___________________________ Which species is the simplest? ________ How ...
... What would it mean if a species’ line stopped before it got to the top? Which two species on the branching tree are the most closely related (have the most in common with each other)? Which two species have the least in common? ___________________________ Which species is the simplest? ________ How ...
CSCE590/822 Data Mining Principles and Applications
... Neighbor-joining produces an unrooted tree How do we root a tree between N species using n-j? An outgroup is a species that we know to be more distantly related to all remaining species, than they are to one another Example: Human, mouse, rat, pig, dog, chicken, whale ...
... Neighbor-joining produces an unrooted tree How do we root a tree between N species using n-j? An outgroup is a species that we know to be more distantly related to all remaining species, than they are to one another Example: Human, mouse, rat, pig, dog, chicken, whale ...
Machine Learning in Computational Biology CSC 2431
... space – if there is signal in a combination of features, it’ll be lost Focusing on consensus – what if there is complementary information? ...
... space – if there is signal in a combination of features, it’ll be lost Focusing on consensus – what if there is complementary information? ...
Estimating cancer survival and clinical outcome based on genetic
... Determines the progression status of human tumors They are defined for tumor samples that are represented by binary vectors indicating the occurrence of a list of genetic events(x1,…,xl) ...
... Determines the progression status of human tumors They are defined for tumor samples that are represented by binary vectors indicating the occurrence of a list of genetic events(x1,…,xl) ...