Raw copy number (CN) data
... • Goal:To partition the clones into sets with the same copy number and to characterize the genomic segments. Noise reduction Detection of Loss, Normal, Gain, Amplification Breakpoint analysis • Biological model: genomic rearrangements lead to gains or losses of sizable contiguous parts of the ...
... • Goal:To partition the clones into sets with the same copy number and to characterize the genomic segments. Noise reduction Detection of Loss, Normal, Gain, Amplification Breakpoint analysis • Biological model: genomic rearrangements lead to gains or losses of sizable contiguous parts of the ...
Clustering – Exercises
... These images give you a view to the distance matrix even without the dendrogram. If you look at the image generated from samples, you’ll notice that there are some clusters of highly correlated samples, mostly near the diagonal line running from lower left-hand corner to the upper right-hand corner. ...
... These images give you a view to the distance matrix even without the dendrogram. If you look at the image generated from samples, you’ll notice that there are some clusters of highly correlated samples, mostly near the diagonal line running from lower left-hand corner to the upper right-hand corner. ...
GENETICS OF CONTINUOUS VARIATION
... flat contradiction to the Mendelian scheme. In 1906 Yule showed that Pearson’s conclusions rested on the specific assumption of complete dominance for all pairs of genes concerned, and that if dominance was sometimes incomplete, the Mendelian scheme could give correlations throughout the actually ob ...
... flat contradiction to the Mendelian scheme. In 1906 Yule showed that Pearson’s conclusions rested on the specific assumption of complete dominance for all pairs of genes concerned, and that if dominance was sometimes incomplete, the Mendelian scheme could give correlations throughout the actually ob ...
Comparative Anatomy: Phylogenetics Assignment
... For this part of the assignment, you must complete the final analyses of our phylogenetic analysis of the Actinopterygii (the ray-finned fishes). While we have one evolutionary hypothesis of the phylogenetic relationships among the fishes we examined by morphological analysis (MacClade parsimony tre ...
... For this part of the assignment, you must complete the final analyses of our phylogenetic analysis of the Actinopterygii (the ray-finned fishes). While we have one evolutionary hypothesis of the phylogenetic relationships among the fishes we examined by morphological analysis (MacClade parsimony tre ...
... analysis is a powerful methodology to investigate not only history but also selection mechanisms and function of biological networks at all levels [1]. Phylogenetic trees based on single loci should be viewed with caution and the best approach is to examine numerous loci across the genome. Due to la ...
Experimental Design
... Compare the number of FDR significant genes What is the overlap? design2 <- model.matrix(~exp.num+strain) The p.value matrix will have four columns instead of 2. Note that the blocking variables should be first in the formula if any kind of anova will be done. ...
... Compare the number of FDR significant genes What is the overlap? design2 <- model.matrix(~exp.num+strain) The p.value matrix will have four columns instead of 2. Note that the blocking variables should be first in the formula if any kind of anova will be done. ...
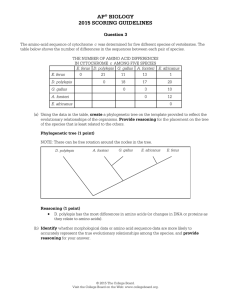
ap® biology 2015 scoring guidelines
... This question focused on using evidence to support biological evolution. Students were asked to evaluate amino acid sequences from five related species to construct a phylogenetic tree reflecting the evolutionary relationships among them, and justify the placement on the tree of the species that is ...
... This question focused on using evidence to support biological evolution. Students were asked to evaluate amino acid sequences from five related species to construct a phylogenetic tree reflecting the evolutionary relationships among them, and justify the placement on the tree of the species that is ...
Ohio State Talk, October 2004
... biased: cases are vastly over-represented • Ordinary logistic regression computes the probability of disease given the environment, given the gene, and given that the person was selected into the case control study ...
... biased: cases are vastly over-represented • Ordinary logistic regression computes the probability of disease given the environment, given the gene, and given that the person was selected into the case control study ...
Linking Genotypes and Phenotypes Peter J. Park, PhD
... •The usual paradigm in a clinical study is having few variables and many samples •Many statistical methods may not be valid without modifications; methods need to be applied with caution ...
... •The usual paradigm in a clinical study is having few variables and many samples •Many statistical methods may not be valid without modifications; methods need to be applied with caution ...
Fine scale mapping
... taking the value 1 if b has a parent in the gene tree and 0 otherwise. Allows for singleton leaf nodes, corresponding to sporadic case chromosomes, and disconnected sub-trees, corresponding to independent mutation events at the same disease locus. Assume number of branches of gene tree not removed i ...
... taking the value 1 if b has a parent in the gene tree and 0 otherwise. Allows for singleton leaf nodes, corresponding to sporadic case chromosomes, and disconnected sub-trees, corresponding to independent mutation events at the same disease locus. Assume number of branches of gene tree not removed i ...
Constraint-Techniques for Collaborative Design
... • Standard errors are determine per expression value Claudio Lottaz: BioConductor - R for Microarray Analysis ...
... • Standard errors are determine per expression value Claudio Lottaz: BioConductor - R for Microarray Analysis ...
Microarray Analysis 3
... clusters there should be? How strict do I want to be? Spilt or Join? Can a gene be in multiple clusters? Hard or soft boundaries between clusters ...
... clusters there should be? How strict do I want to be? Spilt or Join? Can a gene be in multiple clusters? Hard or soft boundaries between clusters ...
AGO1-IP approach to small RNA target discovery in Arabidopsis
... nature of this mutation, as null alleles are embryonic-lethal. Use of conditional, as opposed to constitutive, expression of VSRs is thus an anticipated refinement of the method that might lead to much more tractable effects on target gene accumulation. The scope of the method might also be further ...
... nature of this mutation, as null alleles are embryonic-lethal. Use of conditional, as opposed to constitutive, expression of VSRs is thus an anticipated refinement of the method that might lead to much more tractable effects on target gene accumulation. The scope of the method might also be further ...
Text S1.
... half of the S-genes are known. This is promising as we can expect the best possible performance of the methods even if we use only half of the known underlying network. Conversely, all methods’ performances were low when smaller proportions of known Sgenes were used, especially at 20-33% (3-5 S-gene ...
... half of the S-genes are known. This is promising as we can expect the best possible performance of the methods even if we use only half of the known underlying network. Conversely, all methods’ performances were low when smaller proportions of known Sgenes were used, especially at 20-33% (3-5 S-gene ...
Recursive partitioning for tumor classification with gene
... Results From Classification Tree on the Data Fig 1. Classification tree for tissue types by using expression data from three genes ( M26383, R15447, M28214) ...
... Results From Classification Tree on the Data Fig 1. Classification tree for tissue types by using expression data from three genes ( M26383, R15447, M28214) ...
PowerPoint
... 2. Parsimony – fewest number of evolutionary events (mutations) – relatively often fails to reconstruct correct phylogeny, but methods have improved recently 3. Maximum likelihood – L = Pr[Data|Tree] – most flexible class of methods - user-specified evolutionary methods can be used ...
... 2. Parsimony – fewest number of evolutionary events (mutations) – relatively often fails to reconstruct correct phylogeny, but methods have improved recently 3. Maximum likelihood – L = Pr[Data|Tree] – most flexible class of methods - user-specified evolutionary methods can be used ...
Additional File 1
... late stage=1) was assigned for arrays and checked for any MEs if they were significantly correlated with that indicator. Modules that significantly correlate with a stage phenotype were labeled “candidate modules”. In our analysis, only one module (pink) was found to correlate with a stage and there ...
... late stage=1) was assigned for arrays and checked for any MEs if they were significantly correlated with that indicator. Modules that significantly correlate with a stage phenotype were labeled “candidate modules”. In our analysis, only one module (pink) was found to correlate with a stage and there ...
A PCA Based Method of Gene Expression Visual Analysis
... ponents of this axis are calculated using principal components. For (Fig. 2). example, in case of Fig. 2, genes that contribute to divide groups are calculated and users get a gene list. The last process of our proposal is using chromosomal viewer (Fig. 5). Genes include the gene list are distribute ...
... ponents of this axis are calculated using principal components. For (Fig. 2). example, in case of Fig. 2, genes that contribute to divide groups are calculated and users get a gene list. The last process of our proposal is using chromosomal viewer (Fig. 5). Genes include the gene list are distribute ...
FILTUS: a desktop GUI for fast and efficient
... existing programs which are limited to Variant Call Format (VCF) or other specific input formats. Typical examples of non-standard formats are VCF files with additional columns, and files produced by Annovar (Wang et al., 2010). Although FILTUS is primarily intended for WES-scale data, whole-genome ...
... existing programs which are limited to Variant Call Format (VCF) or other specific input formats. Typical examples of non-standard formats are VCF files with additional columns, and files produced by Annovar (Wang et al., 2010). Although FILTUS is primarily intended for WES-scale data, whole-genome ...
Computational Biology
... It has been suggested that nucleotides within a given gene do not evolve independently. Re-sample subset of orthologous nucleotides from the total data set. Only 3000 randomly chosen nucleotide positions (corresponding to less than three concatenated genes) are sufficient to generate single tree wit ...
... It has been suggested that nucleotides within a given gene do not evolve independently. Re-sample subset of orthologous nucleotides from the total data set. Only 3000 randomly chosen nucleotide positions (corresponding to less than three concatenated genes) are sufficient to generate single tree wit ...
PowerPoint
... • Comparative biology is based upon evolutionary relationships between compared entities • Evolutionary relationships are normally depicted in a phylogenetic tree ...
... • Comparative biology is based upon evolutionary relationships between compared entities • Evolutionary relationships are normally depicted in a phylogenetic tree ...
Molecular phylogeny, part B
... Easy alignments: Correctly aligning the homologous sequence is the next task. In some cases it is an easy task. A simple sequence alignment is shown below: ...
... Easy alignments: Correctly aligning the homologous sequence is the next task. In some cases it is an easy task. A simple sequence alignment is shown below: ...
Natural selection and phylogenetic analysis
... workhorse of phylogenetics near the species level (phylogeography) during the 1990s (17), and in recent years whole-mitochondrial genome sequencing has been used to understand the phylogenetic relationships of many groups, especially vertebrates, for which there are now hundreds of complete genomes. ...
... workhorse of phylogenetics near the species level (phylogeography) during the 1990s (17), and in recent years whole-mitochondrial genome sequencing has been used to understand the phylogenetic relationships of many groups, especially vertebrates, for which there are now hundreds of complete genomes. ...