and Light-Chain Variable-Region Gene Families
... VH genes. The Vl sequences from humans belong to several groups, whereas the sequences from chickens, rabbits, cattle, and sheep are restricted to one Vl group. The two remaining species, mice and horses, show a somewhat unusual pattern. In mice, in contrast to numerous VH genes (see fig. 1), there ...
... VH genes. The Vl sequences from humans belong to several groups, whereas the sequences from chickens, rabbits, cattle, and sheep are restricted to one Vl group. The two remaining species, mice and horses, show a somewhat unusual pattern. In mice, in contrast to numerous VH genes (see fig. 1), there ...
Conspiracy of silence among repeated transgenes
... reflects normal influences at the site of insertion. However, genomes have mechanisms for recognizing potentially threatening sequence elements, like transposons, and silencing them. Cytosine methylation is believed to serve this purpose in most eukaryotic taxa.(26) RIGS seems to be another genome d ...
... reflects normal influences at the site of insertion. However, genomes have mechanisms for recognizing potentially threatening sequence elements, like transposons, and silencing them. Cytosine methylation is believed to serve this purpose in most eukaryotic taxa.(26) RIGS seems to be another genome d ...
Gene Expression in Thyroxin-Induced Metamorphosing Axolotl Hearts
... The Mexican axolotl (Ambystoma mexicanum) is a unique model to study vertebrate heart development for several reasons. In addition to the wild-type animal, there is also an embryonic lethal condition caused by a homozygous recessive mutation in cardiac gene “c” [1,2]. These mutant embryonic hearts d ...
... The Mexican axolotl (Ambystoma mexicanum) is a unique model to study vertebrate heart development for several reasons. In addition to the wild-type animal, there is also an embryonic lethal condition caused by a homozygous recessive mutation in cardiac gene “c” [1,2]. These mutant embryonic hearts d ...
187-192. Control of transcription by Pontin and Reptin
... levels. Yeast Uri1 plays a similar role, and it does so by repressing the translation of the transcription factor Gcn4. Thus, the Uri1 complex contributes indirectly to the control of gene expression, but it might also affect transcription more directly since at least Uri1 physically interacts with ...
... levels. Yeast Uri1 plays a similar role, and it does so by repressing the translation of the transcription factor Gcn4. Thus, the Uri1 complex contributes indirectly to the control of gene expression, but it might also affect transcription more directly since at least Uri1 physically interacts with ...
The Arabidopsis NAC Transcription Factor VNI2
... influenced by both developmental and environmental cues. Gene expression analysis using the GENEVESTIGATOR database (https://www.genevestigator.com/gv/index.jsp) revealed that VNI2, which is highly expressed in senescing plant tissues, is also induced by environmental stress conditions, suggesting t ...
... influenced by both developmental and environmental cues. Gene expression analysis using the GENEVESTIGATOR database (https://www.genevestigator.com/gv/index.jsp) revealed that VNI2, which is highly expressed in senescing plant tissues, is also induced by environmental stress conditions, suggesting t ...
1st set of Journal Clubs this Wednesday!
... •Regulatory sequences adjacent to an operon determine whether it’s transcribed or not. •Regulatory sequences are primarily ‘operators’ (repressor binding sequences). Can also include activator binding sequences. •Regulatory proteins work with regulatory sequences to control transcription of the oper ...
... •Regulatory sequences adjacent to an operon determine whether it’s transcribed or not. •Regulatory sequences are primarily ‘operators’ (repressor binding sequences). Can also include activator binding sequences. •Regulatory proteins work with regulatory sequences to control transcription of the oper ...
Imprinted gene expression in hybrids: perturbed
... The developmental establishment and maintenance of imprinting involve complex interactions between cis-regulatory elements and trans-acting factors, some of which are themselves subject to imprinting (Kelsey and Feil, 2013; Wolf, 2013). The transcription factor gene ZAC1 is imprinted in both mice an ...
... The developmental establishment and maintenance of imprinting involve complex interactions between cis-regulatory elements and trans-acting factors, some of which are themselves subject to imprinting (Kelsey and Feil, 2013; Wolf, 2013). The transcription factor gene ZAC1 is imprinted in both mice an ...
Gene density and transcription influence the localization of
... 11p15 contains clusters of imprinted genes. The mechanistic basis of imprinting is yet to be fully defined, but aspects of higher-order chromatin structure have been implicated, and homologous loci of human and mouse genes ...
... 11p15 contains clusters of imprinted genes. The mechanistic basis of imprinting is yet to be fully defined, but aspects of higher-order chromatin structure have been implicated, and homologous loci of human and mouse genes ...
msb201035-sup
... (Fisher exact test, p<0.05). The fact that the timing of Tinman binding is not highly correlation with the timing of its target genes’ expression most likely reflects the importance of co-operativity with other TFs for Tinman function, as previously reported (e.g. HsiuHsiang Lee and Manfred Frasch, ...
... (Fisher exact test, p<0.05). The fact that the timing of Tinman binding is not highly correlation with the timing of its target genes’ expression most likely reflects the importance of co-operativity with other TFs for Tinman function, as previously reported (e.g. HsiuHsiang Lee and Manfred Frasch, ...
Chapter 15 The Chromosomal Basis of Inheritance
... ° This gene is active only on the Barr-body chromosome and produces multiple copies of an RNA molecule that attach to the X chromosome on which they were made. ° This initiates X inactivation. ° The mechanism that connects XIST RNA and DNA methylation is unknown. ...
... ° This gene is active only on the Barr-body chromosome and produces multiple copies of an RNA molecule that attach to the X chromosome on which they were made. ° This initiates X inactivation. ° The mechanism that connects XIST RNA and DNA methylation is unknown. ...
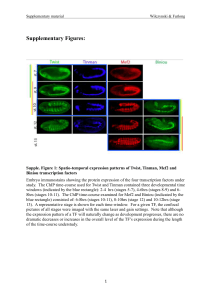
A novel C. elegans zinc finger transcription factor, lsy
... ASEL neuronal fate has switched to an ASER neuronal fate (see Fig. 1B for an example). One mutant, ot71, defines the lsy6 miRNA locus (Johnston and Hobert, 2003); three mutants, ot69, ot74 and ot78, lie within the lin-49 transcription factor locus (Chang et al., 2003); one mutant, ot79, defines the ...
... ASEL neuronal fate has switched to an ASER neuronal fate (see Fig. 1B for an example). One mutant, ot71, defines the lsy6 miRNA locus (Johnston and Hobert, 2003); three mutants, ot69, ot74 and ot78, lie within the lin-49 transcription factor locus (Chang et al., 2003); one mutant, ot79, defines the ...
Chapter 15 – The Chromosomal Basis of Inheritance
... Mary Lyon, a British geneticist, demonstrated that selection of which X chromosome will form the Barr body occurs randomly and independently in embryonic cells at the time of X inactivation. ...
... Mary Lyon, a British geneticist, demonstrated that selection of which X chromosome will form the Barr body occurs randomly and independently in embryonic cells at the time of X inactivation. ...
Evolution of Immunoglobulin Kappa Chain Variable Region
... a sequence similarity of 95%–100%, and this duplication has not been found in the chimpanzee, gorilla, or orangutan (Ermert et al. 1995). From information on the overall sequence divergence between the two sets of duplicate genes (;1%), Schäble and Zachau (1993) suggested that the duplication occur ...
... a sequence similarity of 95%–100%, and this duplication has not been found in the chimpanzee, gorilla, or orangutan (Ermert et al. 1995). From information on the overall sequence divergence between the two sets of duplicate genes (;1%), Schäble and Zachau (1993) suggested that the duplication occur ...
Expression of the six chromate ion transporter
... universal primers. DNA fragments containing the chr genes were obtained by digestions with HindIII/XbaI or HindIII/EcoRI endonucleases and subcloned into the corresponding sites of pACYC184 or pUCP20 vectors. E. coli W3110 cells were transformed by electroporation with recombinant plasmids and trans ...
... universal primers. DNA fragments containing the chr genes were obtained by digestions with HindIII/XbaI or HindIII/EcoRI endonucleases and subcloned into the corresponding sites of pACYC184 or pUCP20 vectors. E. coli W3110 cells were transformed by electroporation with recombinant plasmids and trans ...
bbr038online 474..484 - Oxford Academic
... compared with the traditional Sanger method, genome sequencing based on new technologies will yield more genome sequences that are not well assembled [2, 3], which in turn will hinder comprehensive annotation of the protein-coding landscape. In particular, a major challenge is the identification of ...
... compared with the traditional Sanger method, genome sequencing based on new technologies will yield more genome sequences that are not well assembled [2, 3], which in turn will hinder comprehensive annotation of the protein-coding landscape. In particular, a major challenge is the identification of ...
DNA Chips: Genes to Disease
... First, you will prepare your DNA microarray by spotting each of the six different gene sequences onto a glass slide. For real microarrays, scientists actually print thousands of microscopic DNA spots onto a slide, one spot for each gene they want to examine. Your spots will be much larger than those ...
... First, you will prepare your DNA microarray by spotting each of the six different gene sequences onto a glass slide. For real microarrays, scientists actually print thousands of microscopic DNA spots onto a slide, one spot for each gene they want to examine. Your spots will be much larger than those ...
An In Silico Investigation Into the Discovery of Novel Cis
... The intronic regions of the PAX7 gene were analyzed using computational DNA pattern recognition methods. Several potential cis-regulatory motifs were identified in this investigation and one in particular that was common to both PAX7 and PAX3 and also to NF1, could have implications for the role of ...
... The intronic regions of the PAX7 gene were analyzed using computational DNA pattern recognition methods. Several potential cis-regulatory motifs were identified in this investigation and one in particular that was common to both PAX7 and PAX3 and also to NF1, could have implications for the role of ...
Turning floral organs into leaves, leaves into floral organs Koji Goto
... with the proposed role of promoting the growth of marginal tissues [22••]. SPT is also expressed in many different tissues outside the carpel, suggesting that it may act redundantly in diverse developmental processes. The negative regulation of SPT by the A class genes is at the transcriptional leve ...
... with the proposed role of promoting the growth of marginal tissues [22••]. SPT is also expressed in many different tissues outside the carpel, suggesting that it may act redundantly in diverse developmental processes. The negative regulation of SPT by the A class genes is at the transcriptional leve ...
Reveal—visual eQTL analytics
... information) with specific phenotypic outcomes. Based on this aggregated view of the data, analysts (e.g. biologists and bioinformaticians) are enabled to infer causal associations of genetic variation, expression and disease, i.e. to identify the genetic basis for phenotypic variations. More genera ...
... information) with specific phenotypic outcomes. Based on this aggregated view of the data, analysts (e.g. biologists and bioinformaticians) are enabled to infer causal associations of genetic variation, expression and disease, i.e. to identify the genetic basis for phenotypic variations. More genera ...
Document
... • hypothesis: autosomally-encoded blocking factor binds to the Xa chromosome and prevents its inactivation – sequence known as the XIC – X inactivation center – may bind these binding factors and prevent inactivation?? • the XIC of the the Xi chromosome produces a non-coding RNA called Xist RNA – co ...
... • hypothesis: autosomally-encoded blocking factor binds to the Xa chromosome and prevents its inactivation – sequence known as the XIC – X inactivation center – may bind these binding factors and prevent inactivation?? • the XIC of the the Xi chromosome produces a non-coding RNA called Xist RNA – co ...
The evolution of developmental gene networks
... identify—through comparative analysis of developmental data within a phylogenetic framework—the changes in developmental mechanisms that underpinned divergence in body architecture between lineages. Rather than thinking in terms of developmental time—with the rather arbitrary starting point of zygot ...
... identify—through comparative analysis of developmental data within a phylogenetic framework—the changes in developmental mechanisms that underpinned divergence in body architecture between lineages. Rather than thinking in terms of developmental time—with the rather arbitrary starting point of zygot ...
1 Oviduct-embryo interactions in cattle
... Copyright 2015 by The Society for the Study of Reproduction. ...
... Copyright 2015 by The Society for the Study of Reproduction. ...