Protein Structure and Folding
... Protein Structure and Folding Exercise 2 by 27.2.2002 1. Many proteins in particular small ones are stabilized by disulfide bridges. Lysozyme contains 8 cysteine residues that form 4 four bridges. Derive expression for the number of ways N cysteins can pair with each other. Some proteins like many n ...
... Protein Structure and Folding Exercise 2 by 27.2.2002 1. Many proteins in particular small ones are stabilized by disulfide bridges. Lysozyme contains 8 cysteine residues that form 4 four bridges. Derive expression for the number of ways N cysteins can pair with each other. Some proteins like many n ...
Introduction to Protein Science Architecture, Function
... Ex) Sickle cell anaemia, Z-mutant of a1-antitrypsin ...
... Ex) Sickle cell anaemia, Z-mutant of a1-antitrypsin ...
E U F T DG Unfolded state, ensemble Native fold, one
... Explain why some backbone conformations are favoured and some are “forbidden” (not found in natural proteins). – Name properties on which the amino acids can be grouped. – Explain the driving forces behind protein folding related to the properties of the backbone and the side chains. ...
... Explain why some backbone conformations are favoured and some are “forbidden” (not found in natural proteins). – Name properties on which the amino acids can be grouped. – Explain the driving forces behind protein folding related to the properties of the backbone and the side chains. ...
Test 2 - HCC Learning Web
... 3. Polysaccharides, triacylglycerides, and proteins are similar. Explain? 4. Stanley Miller's 1953 experiments proved that. Explain? 5. Why are hydrocarbons insoluble in water? 6. Humans can digest starch but not cellulose because. Explain? 7. How does RNA differ from DNA? 8. Explain how ATP functio ...
... 3. Polysaccharides, triacylglycerides, and proteins are similar. Explain? 4. Stanley Miller's 1953 experiments proved that. Explain? 5. Why are hydrocarbons insoluble in water? 6. Humans can digest starch but not cellulose because. Explain? 7. How does RNA differ from DNA? 8. Explain how ATP functio ...
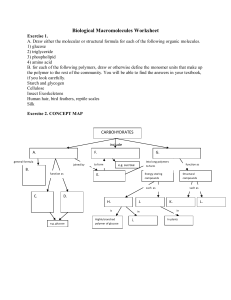
Biological Macromolecules Worksheet
... a. the number _____ of different nitrogenous bases in DNA b. the number _____ of different chemical classes of amino acids c. the number _____ of chains of nucleotides in a DNA molecule d. the number _____ of different nitrogenous bases in RNA e. the number _____ of different amino acids found in pr ...
... a. the number _____ of different nitrogenous bases in DNA b. the number _____ of different chemical classes of amino acids c. the number _____ of chains of nucleotides in a DNA molecule d. the number _____ of different nitrogenous bases in RNA e. the number _____ of different amino acids found in pr ...
OriGene Technologies launches over 5,000 heavy isotope labeled
... academic, pharmaceutical and biotech companies in their research of gene functions and drug discovery. OriGene develops proteins, antibodies, and other molecular tools to allow researchers to analyze their data on a multiplex level. OriGene's novel product line includes the world's largest cDNA and ...
... academic, pharmaceutical and biotech companies in their research of gene functions and drug discovery. OriGene develops proteins, antibodies, and other molecular tools to allow researchers to analyze their data on a multiplex level. OriGene's novel product line includes the world's largest cDNA and ...
Teaching Notes
... 3. Small globular proteins (made of one single polymer chain) present in the cytoplasm or secreted by the cell, have their hydrophobic amino acid residues tucked in the core of the protein. 4. Larger protein complexes (formed from multiple copies of the same or different proteins) may have protein c ...
... 3. Small globular proteins (made of one single polymer chain) present in the cytoplasm or secreted by the cell, have their hydrophobic amino acid residues tucked in the core of the protein. 4. Larger protein complexes (formed from multiple copies of the same or different proteins) may have protein c ...
Proteins
... 6. Label the type of bond used to make proteins. 7. Draw arrows to identify these bonds in your model 8. Label the N-terminus and C-terminus 9. Put SQUARES around the R groups 10. Use your amino acid chart to identify & label the type of R group (non-polar, polar, charge basic, charged acidic, etc) ...
... 6. Label the type of bond used to make proteins. 7. Draw arrows to identify these bonds in your model 8. Label the N-terminus and C-terminus 9. Put SQUARES around the R groups 10. Use your amino acid chart to identify & label the type of R group (non-polar, polar, charge basic, charged acidic, etc) ...
Detecting topological patterns in protein networks
... Yeast two-hybrid technique uses two “hybrid proteins”: bait X* (X fused with Gal4p DNA-binding domain) and prey Y* (Y fused with Gal4p activation domain) • Cons: wrong (very high) concentrations, localization (unless both proteins are nuclear), and even host organism (unless done in yeast) • Pros: ...
... Yeast two-hybrid technique uses two “hybrid proteins”: bait X* (X fused with Gal4p DNA-binding domain) and prey Y* (Y fused with Gal4p activation domain) • Cons: wrong (very high) concentrations, localization (unless both proteins are nuclear), and even host organism (unless done in yeast) • Pros: ...
Dynamic visualization of protein molecules in action by highspeed
... techniques has precluded it. Even with the recently emerging super-resolution fluorescence microscopes, this dream is unachievable because they are indirect imaging techniques. Atomic force microscopy (AFM) is a versatile technique to directly image proteins in liquids at submolecular resolution. Ho ...
... techniques has precluded it. Even with the recently emerging super-resolution fluorescence microscopes, this dream is unachievable because they are indirect imaging techniques. Atomic force microscopy (AFM) is a versatile technique to directly image proteins in liquids at submolecular resolution. Ho ...
2.4 review
... 2) Draw a condensation reaction between two amino acids. What is the name of the bond that is formed as a result? 3) Discuss why the same 20 amino organisms are used by most organisms to make proteins. 4) Distinguish between a polypeptide and a protein. 5) What is an “R” group? How many different on ...
... 2) Draw a condensation reaction between two amino acids. What is the name of the bond that is formed as a result? 3) Discuss why the same 20 amino organisms are used by most organisms to make proteins. 4) Distinguish between a polypeptide and a protein. 5) What is an “R” group? How many different on ...
Folding in the cell Cytosolic proteins
... The structure of proteins is determined by the amino acid sequence; many proteins in solution can be unfolded by heat and other denaturants such as high concentrations of urea and guanidinium chloride, but they will spontaneously refold on returning conditions to normal. This refolding takes place i ...
... The structure of proteins is determined by the amino acid sequence; many proteins in solution can be unfolded by heat and other denaturants such as high concentrations of urea and guanidinium chloride, but they will spontaneously refold on returning conditions to normal. This refolding takes place i ...
Protein Folding and Membrane Structure
... • Serve as substrates for biochemical and signaling reactions ...
... • Serve as substrates for biochemical and signaling reactions ...
Flexibility of a polypeptide chain
... ~3 residues/turn, 3 helices wind in a superhelical cable that is stabilized by H-bond in between strands (Pro-OH participates in H-bonding network and lack of –OH on Pro in collagen lead to the disease scurvy (Vitamin C deficiency, ascorbate reduces Fe3+ to Fe2+ in prolyl hydroxylase for its ...
... ~3 residues/turn, 3 helices wind in a superhelical cable that is stabilized by H-bond in between strands (Pro-OH participates in H-bonding network and lack of –OH on Pro in collagen lead to the disease scurvy (Vitamin C deficiency, ascorbate reduces Fe3+ to Fe2+ in prolyl hydroxylase for its ...
Protein Misfolding and Degenerative Diseases
... increases. Why is this? As incredible as it might sound, these diseases are caused not by bacteria or viruses but rather by something conceptually quite simple: incorrect protein folding. Introductory biology courses teach us that proteins are essential for the organism because they participate in v ...
... increases. Why is this? As incredible as it might sound, these diseases are caused not by bacteria or viruses but rather by something conceptually quite simple: incorrect protein folding. Introductory biology courses teach us that proteins are essential for the organism because they participate in v ...
IN THIS ISSUE Mutating it all Discovering ubiquitylation
... Expressed protein ligation (EPL) is a powerful tool for synthetically generating proteins with desired posttranslational modifications. In EPL, one portion of a target protein is expressed in cells as a fusion to an intein, which cleaves itself off, leaving a C-terminal thioester. This thioester can ...
... Expressed protein ligation (EPL) is a powerful tool for synthetically generating proteins with desired posttranslational modifications. In EPL, one portion of a target protein is expressed in cells as a fusion to an intein, which cleaves itself off, leaving a C-terminal thioester. This thioester can ...
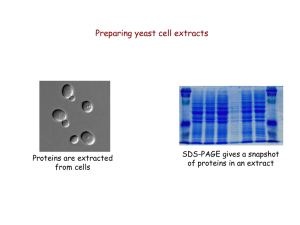
Power Point
... “Quick and dirty” protein extraction from yeast 1. Collect cells by centrifugation 2. Wash cells with deionized water ...
... “Quick and dirty” protein extraction from yeast 1. Collect cells by centrifugation 2. Wash cells with deionized water ...
Shin-ichi Tate Research Group Activity ・ Protein dynamics and
... ・ Protein dynamics and function relationships revealed through nuclear spin relaxation analyses Protein dynamics, in the time regime in sec-msec, can be revealed by nuclear spin relaxations. Systematic analyses on the dynamical modulations caused by single site-directed mutation will give us experi ...
... ・ Protein dynamics and function relationships revealed through nuclear spin relaxation analyses Protein dynamics, in the time regime in sec-msec, can be revealed by nuclear spin relaxations. Systematic analyses on the dynamical modulations caused by single site-directed mutation will give us experi ...
7.5 Proteins – summary of mark schemes
... F. tertiary structure / level: 3-dimensional conformation of a polypeptide / protein; G. held with ionic bonds, hydrogen bonds, disulfide bonds / bridges and hydrophobic bonds; (must give at least two bonds) H. determines overall shape / a named example eg: active sites on enzymes; I. J. K. L. ...
... F. tertiary structure / level: 3-dimensional conformation of a polypeptide / protein; G. held with ionic bonds, hydrogen bonds, disulfide bonds / bridges and hydrophobic bonds; (must give at least two bonds) H. determines overall shape / a named example eg: active sites on enzymes; I. J. K. L. ...
Proteomics Problem Set Lecture 11, CH908 Mass Spectrometry
... 3 (A) Using NCBI protein database ( http://www.ncbi.nlm.nih.gov/protein/ ) and MS-Digest tool (Protein Prospector), perform in-silico digest for the following proteins: murine myoglobin, human myoglobin, chicken ovalbumin, bovine serum albumin, human fibrinogen. Consider oxidation of methionin as a ...
... 3 (A) Using NCBI protein database ( http://www.ncbi.nlm.nih.gov/protein/ ) and MS-Digest tool (Protein Prospector), perform in-silico digest for the following proteins: murine myoglobin, human myoglobin, chicken ovalbumin, bovine serum albumin, human fibrinogen. Consider oxidation of methionin as a ...
View attached file
... Daniel Segal - Research 'Conformational diseases' are diseases caused by misfolding of a protein, often as a result of a missense mutation that does not necessarily disrupt the active site of the protein. As a result, the protein may lose its function, and often the misfolded monomers self-assemble ...
... Daniel Segal - Research 'Conformational diseases' are diseases caused by misfolding of a protein, often as a result of a missense mutation that does not necessarily disrupt the active site of the protein. As a result, the protein may lose its function, and often the misfolded monomers self-assemble ...
TIGR_ISS
... Visually inspect alignments, look for conserved active sites, look for (generally) at least 35% identity across the full lengths of both proteins. If matches are not full length, look to see if there are recognized functional domains in the area where the match occurs. Decide how much information ca ...
... Visually inspect alignments, look for conserved active sites, look for (generally) at least 35% identity across the full lengths of both proteins. If matches are not full length, look to see if there are recognized functional domains in the area where the match occurs. Decide how much information ca ...
Protein–protein interaction

Protein–protein interactions (PPIs) refer to physical contacts established between two or more proteins as a result of biochemical events and/or electrostatic forces.In fact, proteins are vital macromolecules, at both cellular and systemic levels, but they rarely act alone. Diverse essential molecular processes within a cell are carried out by molecular machines that are built from a large number of protein components organized by their PPIs. Indeed, these interactions are at the core of the entire interactomics system of any living cell and so, unsurprisingly, aberrant PPIs are on the basis of multiple diseases, such as Creutzfeld-Jacob, Alzheimer's disease, and cancer.PPIs have been studied from different perspectives: biochemistry, quantum chemistry, molecular dynamics, signal transduction, among others. All this information enables the creation of large protein interaction networks – similar to metabolic or genetic/epigenetic networks – that empower the current knowledge on biochemical cascades and disease pathogenesis, as well as provide putative new therapeutic targets.