CH 107 SI Summer 2015 Worksheet 13 Answers What are the two
... α-helices and β-sheets Hydrogen bonds 2. What types of interactions can be present in tertiary protein structure? Rank the interactions from strongest to weakest. disulfide bonds >> salt bridges > hydrogen bonds > hydrophilic interactions > hydrophobic interactions 3. Compare and contrast globular a ...
... α-helices and β-sheets Hydrogen bonds 2. What types of interactions can be present in tertiary protein structure? Rank the interactions from strongest to weakest. disulfide bonds >> salt bridges > hydrogen bonds > hydrophilic interactions > hydrophobic interactions 3. Compare and contrast globular a ...
blumberg-lab.bio.uci.edu
... The positives of the approach ● For genome wide interaction mapping this is the most feasible approach. ● Hypothetical networks arise which become of interest for studies. ● Better understanding of cellular processes can be developed by testable hypothesis of these ...
... The positives of the approach ● For genome wide interaction mapping this is the most feasible approach. ● Hypothetical networks arise which become of interest for studies. ● Better understanding of cellular processes can be developed by testable hypothesis of these ...
Overview
... Yuan Lecture 2, Class 24: Protein Folding and Molecular Chaperones April 20th, 2017 Overview The intracellular concentration of protein in bacterial cells can be estimated to be ~135 mg/ml. In this session, we will explore how bacteria employ a suite of molecular machines collectively known as chape ...
... Yuan Lecture 2, Class 24: Protein Folding and Molecular Chaperones April 20th, 2017 Overview The intracellular concentration of protein in bacterial cells can be estimated to be ~135 mg/ml. In this session, we will explore how bacteria employ a suite of molecular machines collectively known as chape ...
Ch 4 Reading Guide
... 3. Give a chemical explanation of why a peptide bond is planar. 4. Why are almost all peptide bonds in proteins trans rather than cis. 5. Regular, folded segments of amino acids near one another in linear sequence is called ____________________ structure. 6. In the -helix, a _____________________ b ...
... 3. Give a chemical explanation of why a peptide bond is planar. 4. Why are almost all peptide bonds in proteins trans rather than cis. 5. Regular, folded segments of amino acids near one another in linear sequence is called ____________________ structure. 6. In the -helix, a _____________________ b ...
analysis of a local huntington protein interaction network
... Huntington's Disease is a neurodegenerative disorder caused by an abnormally long stretch of glutamines in the associated huntingtin protein. This study sheds light on possible functions for the huntingtin protein though analysis of a local protein-protein interaction network consisting of the hunti ...
... Huntington's Disease is a neurodegenerative disorder caused by an abnormally long stretch of glutamines in the associated huntingtin protein. This study sheds light on possible functions for the huntingtin protein though analysis of a local protein-protein interaction network consisting of the hunti ...
Improving Function Prediction Using Patterns of Native Disorder in
... Instrinsically unstructured (disordered) proteins adopt little or no stable secondary structure in their native state. Proteins containing long disordered regions are abundant within eukaryotic genomes and can be predicted successfully from amino sequence. Disordered regions have been shown to be im ...
... Instrinsically unstructured (disordered) proteins adopt little or no stable secondary structure in their native state. Proteins containing long disordered regions are abundant within eukaryotic genomes and can be predicted successfully from amino sequence. Disordered regions have been shown to be im ...
Slide 1 - AccessMedicine
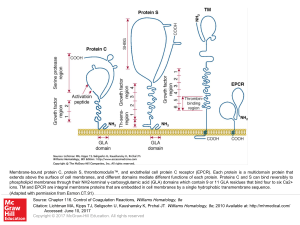
... Membrane-bound protein C, protein S, thrombomodulin™, and endothelial cell protein C receptor (EPCR). Each protein is a multidomain protein that extends above the surface of cell membranes, and different domains mediate different functions of each protein. Proteins C and S can bind reversibly to pho ...
... Membrane-bound protein C, protein S, thrombomodulin™, and endothelial cell protein C receptor (EPCR). Each protein is a multidomain protein that extends above the surface of cell membranes, and different domains mediate different functions of each protein. Proteins C and S can bind reversibly to pho ...
ProteinChipâ technology is one of the most exciting advancements
... last 5 years. The Protein Biology SystemTM (PBS) combines the power of mass analysis with chromatography surfaces on an integrated platform. The PBS can easily be used by biologists, biochemists, and clinicians. ProteinChip uses a wide variety of surface chemistries (normal phase, reverse phase, io ...
... last 5 years. The Protein Biology SystemTM (PBS) combines the power of mass analysis with chromatography surfaces on an integrated platform. The PBS can easily be used by biologists, biochemists, and clinicians. ProteinChip uses a wide variety of surface chemistries (normal phase, reverse phase, io ...
Proteomics
... If the two proteins interact, the reporter gene (here: HIS3) is switched on and the diploids can grow on -His plates: ...
... If the two proteins interact, the reporter gene (here: HIS3) is switched on and the diploids can grow on -His plates: ...
Capturing denaturing proteins * Small Heat Shock Protein substrate
... The ubiquitous small heat shock proteins (sHSPs) act as molecular chaperones to prevent irreversible protein aggregation and are significant components of the protein quality control network. Expression and/or mutation of sHSPs are linked to multiple diseases of protein misfolding, including neurode ...
... The ubiquitous small heat shock proteins (sHSPs) act as molecular chaperones to prevent irreversible protein aggregation and are significant components of the protein quality control network. Expression and/or mutation of sHSPs are linked to multiple diseases of protein misfolding, including neurode ...
Interaction Site Evolution
... COMPUTER SCIENCE - Interaction Site Evolution DNA is the blueprint for generating strings of amino acids which fold into proteins. The interactions these proteins form with each other are primary components of organismal physiology. Proteins assume very specific shapes, and the amino acids on their ...
... COMPUTER SCIENCE - Interaction Site Evolution DNA is the blueprint for generating strings of amino acids which fold into proteins. The interactions these proteins form with each other are primary components of organismal physiology. Proteins assume very specific shapes, and the amino acids on their ...
Abstract
... protein 3D structure, interactions and recognition in signaling networks. Modern sequencing technologies provide us with a rich source of data about the evolutionary history of proteins. Inferring a joint probability distribution of amino acid sequences that are members of a protein family, signals ...
... protein 3D structure, interactions and recognition in signaling networks. Modern sequencing technologies provide us with a rich source of data about the evolutionary history of proteins. Inferring a joint probability distribution of amino acid sequences that are members of a protein family, signals ...
Protein interactions are essential for many biological functions to occur. ... Erika Lacy: Cell Biology & Neuroscience
... Fluorescent Probes for Detecting Protein Interactions in Bacteria Protein interactions are essential for many biological functions to occur. Bimolecular Fluorescence Complementation (BiFC) assay is a complementation-based technique used to study protein interactions. One benefit of this approach is ...
... Fluorescent Probes for Detecting Protein Interactions in Bacteria Protein interactions are essential for many biological functions to occur. Bimolecular Fluorescence Complementation (BiFC) assay is a complementation-based technique used to study protein interactions. One benefit of this approach is ...
Protein–protein interaction

Protein–protein interactions (PPIs) refer to physical contacts established between two or more proteins as a result of biochemical events and/or electrostatic forces.In fact, proteins are vital macromolecules, at both cellular and systemic levels, but they rarely act alone. Diverse essential molecular processes within a cell are carried out by molecular machines that are built from a large number of protein components organized by their PPIs. Indeed, these interactions are at the core of the entire interactomics system of any living cell and so, unsurprisingly, aberrant PPIs are on the basis of multiple diseases, such as Creutzfeld-Jacob, Alzheimer's disease, and cancer.PPIs have been studied from different perspectives: biochemistry, quantum chemistry, molecular dynamics, signal transduction, among others. All this information enables the creation of large protein interaction networks – similar to metabolic or genetic/epigenetic networks – that empower the current knowledge on biochemical cascades and disease pathogenesis, as well as provide putative new therapeutic targets.