ppt
... Given a multiple alignment (of protein coding DNA) we can convert the DNA to proteins. We can then try to model the phylogenetic relations between the proteins using a fixed rate matrix Q, some phylogeney T and branch lengths ti When modeling hundreds/thousands amino acid sequences, we cannot learn ...
... Given a multiple alignment (of protein coding DNA) we can convert the DNA to proteins. We can then try to model the phylogenetic relations between the proteins using a fixed rate matrix Q, some phylogeney T and branch lengths ti When modeling hundreds/thousands amino acid sequences, we cannot learn ...
Molecular evolution and phylogenetic implications in clinical research
... length of branches. There are two crucial distance-based methods: Unweighted Pair Group Method with Arithmetic Mean (UPGMA) and Neighbour Joining (NJ) method [7]. UPGMA is one of the simplest methods of phylogenetic trees construction. It has numerous limitations and wrong assumptions; therefore, th ...
... length of branches. There are two crucial distance-based methods: Unweighted Pair Group Method with Arithmetic Mean (UPGMA) and Neighbour Joining (NJ) method [7]. UPGMA is one of the simplest methods of phylogenetic trees construction. It has numerous limitations and wrong assumptions; therefore, th ...
Different Data Structures
... A B-tree T is a rooted tree (whose root is T.root) having the properties: Every node x has the following attributes: – x.n, the number of keys currently stored in node x, – the x.n keys themselves, x.key1, x.key2, …, x.keyx.n, stored in nondecreasing order, so that x.key1 <= x.key2 <= … <= x.keyx.n ...
... A B-tree T is a rooted tree (whose root is T.root) having the properties: Every node x has the following attributes: – x.n, the number of keys currently stored in node x, – the x.n keys themselves, x.key1, x.key2, …, x.keyx.n, stored in nondecreasing order, so that x.key1 <= x.key2 <= … <= x.keyx.n ...
Two-point Linkage Analysis: a brief outline of theory
... proportions, but also the specific observations in this family. The sample investigated has been shown to consist of three types of families: in 48% of families, the gene is located on chromosome 8, in 24% of them on chromosome 19, and in 28% of families the gene is located on chromosome 11. There ...
... proportions, but also the specific observations in this family. The sample investigated has been shown to consist of three types of families: in 48% of families, the gene is located on chromosome 8, in 24% of them on chromosome 19, and in 28% of families the gene is located on chromosome 11. There ...
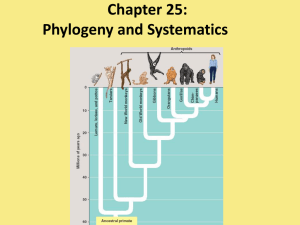
Phylogeny
... What you need to know! The taxonomic categories and how they indicate relatedness. How systematics is used to develop phylogenetic trees. The three domains of life including their similarities and their differences. ...
... What you need to know! The taxonomic categories and how they indicate relatedness. How systematics is used to develop phylogenetic trees. The three domains of life including their similarities and their differences. ...
Objective 2.0
... across the United States with new tools to enhance and accelerate traditional tree improvement activities. These “knowledge-based” tools derive value from experimentally demonstrated associations between traits of interest, like wood density or disease resistance, and the tree’s genetic code (geneti ...
... across the United States with new tools to enhance and accelerate traditional tree improvement activities. These “knowledge-based” tools derive value from experimentally demonstrated associations between traits of interest, like wood density or disease resistance, and the tree’s genetic code (geneti ...
Lecture PPT - Carol Lee Lab
... 2. You are a medical researcher working on HIV. A novel strain has appeared in Madison, Wisconsin. To determine which drugs would be most effective in treating this new strain (because different strains are resistant to different drugs), you need to determine its recent evolutionary history. You de ...
... 2. You are a medical researcher working on HIV. A novel strain has appeared in Madison, Wisconsin. To determine which drugs would be most effective in treating this new strain (because different strains are resistant to different drugs), you need to determine its recent evolutionary history. You de ...
1 - Houston ISD
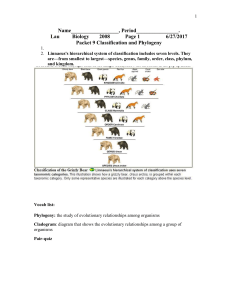
... 2. Linnaeus's hierarchical system of classification includes seven levels. They are—from smallest to largest—species, genus, family, order, class, phylum, and kingdom. ...
... 2. Linnaeus's hierarchical system of classification includes seven levels. They are—from smallest to largest—species, genus, family, order, class, phylum, and kingdom. ...
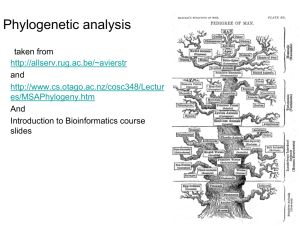
Phylogenetic Analysis
... p-distance: the fraction of non-identical alignment positions (observed distance) ...
... p-distance: the fraction of non-identical alignment positions (observed distance) ...
Chapter 2: The Chemical Context of Life
... appearance (rapid environmental change leads to rapid evolution; also, small changes in genes can lead to large morphological differences) • Organisms that appear similar not always closely related (convergent evolution) • Just because 2 groups share primitive characters does not mean they are close ...
... appearance (rapid environmental change leads to rapid evolution; also, small changes in genes can lead to large morphological differences) • Organisms that appear similar not always closely related (convergent evolution) • Just because 2 groups share primitive characters does not mean they are close ...
Molecular Phylogeny
... ..T.. more closely related to each other than they are to the sequence with G. ...
... ..T.. more closely related to each other than they are to the sequence with G. ...
Introductory Biological Sequence Analysis Through Spreadsheets
... Spreadsheet #3 (cont.) To study the evolution of a sequence, we randomly pick a site for mutation, then change its letter ...
... Spreadsheet #3 (cont.) To study the evolution of a sequence, we randomly pick a site for mutation, then change its letter ...
macroevolutoin part i: phylogenies
... The following rules apply to reconstructing a phylogeny: 1. Maximum likelihood states that when considering multiple phylogenetic hypotheses, one should take into account the one that reflects the most likely sequence of evolutionary events given certain rules about how DNA changes over time. 2. Max ...
... The following rules apply to reconstructing a phylogeny: 1. Maximum likelihood states that when considering multiple phylogenetic hypotheses, one should take into account the one that reflects the most likely sequence of evolutionary events given certain rules about how DNA changes over time. 2. Max ...
PPT - UT Computer Science
... • The state (i.e., nucleotide) of each site at the root is random • The sites evolve independently and identically (i.i.d.) • If the site changes its state on an edge, it changes with equal probability to the other states • For every edge e, p(e) is defined, which is the probability of change for a ...
... • The state (i.e., nucleotide) of each site at the root is random • The sites evolve independently and identically (i.i.d.) • If the site changes its state on an edge, it changes with equal probability to the other states • For every edge e, p(e) is defined, which is the probability of change for a ...
生物計算
... The number of unions is the minimum number of substitutions. For uninformative site, it is the number of different nucleotides minus one. ...
... The number of unions is the minimum number of substitutions. For uninformative site, it is the number of different nucleotides minus one. ...
Comp. Genomics
... • Distinguishes between a target sequence – T and other informative sequences (Is) that may contain gaps • States correspond to sequence types in the target sequence ...
... • Distinguishes between a target sequence – T and other informative sequences (Is) that may contain gaps • States correspond to sequence types in the target sequence ...
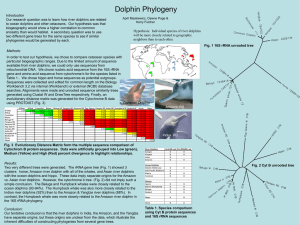
dolphin1
... Our research question was to learn how river dolphins are related to ocean dolphins and other cetaceans. Our hypothesis was that biogeography would show a higher correlation to common ancestry than would habitat. A secondary question was to use two different gene trees for the same species to see if ...
... Our research question was to learn how river dolphins are related to ocean dolphins and other cetaceans. Our hypothesis was that biogeography would show a higher correlation to common ancestry than would habitat. A secondary question was to use two different gene trees for the same species to see if ...
Key terms: Positional homology Homoplasy Reversal Parallelism
... DNA sequences to illustrate these concepts. 2. Assume you have several DNA sequences and that they are sufficiently divergent for multiple substitutions to have occurred at a given site. List and explain the possible sources of homoplasy at such sites. 3. Explain why a phylogenetic concept of homolo ...
... DNA sequences to illustrate these concepts. 2. Assume you have several DNA sequences and that they are sufficiently divergent for multiple substitutions to have occurred at a given site. List and explain the possible sources of homoplasy at such sites. 3. Explain why a phylogenetic concept of homolo ...