* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Lecture 7: Signaling Through Lymphocyte Receptors
Survey
Document related concepts
Transcript
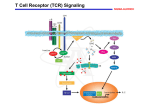
Lecture 7: Signaling Through Lymphocyte Receptors Questions to Consider After recognition of its cognate MHC:peptide, how does the T cell receptor activate immune response genes? What are the structural motifs used by signal transduction molecules that permit specific interaction and activation of effector proteins? How do genes know what is going on in the outside environment requiring them to make their proteins at the appropriate time? General Signal Transduction Concepts Recognition of MHC:Peptide Complex by TCR Initiates Signaling Cascade in the T cell Some Cellular Receptors Have Their Own Transduction Molecules Activated by Dimerization Other Cellular Receptors Are Transduction Molecule-challenged Signal Transduction Pathways Amplify the Initial Signal Signal Transduction Pathways Must Have the Capacity to be Turned Off Adapter Proteins Recruit Target Molecules From the Cytoplasm to Membrane-Associated Kinases Binds to Binds to Binds to P120/130 CD4/CD8 PI3-kinase PI3-kinase (prolineor (pYEEI motif) rich TCR/CD3 motif) Kinase Adapter Adapter Adapter Src-family Kinases Are Regulated Tyrosine Kinases and Also Function as Adapter Proteins Regulates PTK activity PTK (dephosphorylation by Phosphorylation of CD45 upregulates specific tyrosine activity:Phosphorylation by residues within p50csk downregulates TCRz/CD3, CD5 P120/130 Scaffolding Proteins Permit Assembly of Signaling Complexes Overview of T Cell Signal Transduction The T Cell Receptor Complex Consists of Eight Proteins With Unique Functions Ligand binding heterodimer or binds peptide + MHC CD3 Complex and Nonpolymorphic chains present as or dimers Intracellular - present as dimers present as a homodimer Interaction Between MHC Class II:Peptide and TCR Recruits the CD4 Molecule TCR-associated Proteins: src-family Kinases lck binds to CD4 and CD8 fyn binds to non-phosphorylated ITAMs SH3/SH2 domains inactive when bound to phosphorylated C-terminus activated by dephosphorylation by phosphatase such as CD45 Regulation of Src-family Kinases by Phosphorylation and Dephosphorylation of Activation and Inhibitory Tyrosine Sites MHC:Peptide Interaction Recruits the CD45 Tyrosine Phosphatase Which Dephosphorylates and Activates Src-kinases: Fyn and Lck Phosphorylation of Receptors Permits Them to Recruit Adapter Proteins That Bind Phosphotyrosine Residues Role of Immunoreceptor Tyrosine- based Activation Motifs (ITAMs) Tyrosine residues on ITAMs are phosphorylated and thereby bind proteins containing SH2 domains Total of 10 ITAMs/T cell receptor complex 1 ITAM/CD3 or chain = 4 ITAMs 3 ITAMs/CD3 chain = 6 ITAMs Why So Many ITAMs? Required for amplification of signal transduced by few TCR molecules Different transduction molecules bind to different ITAMs Required for association of TCR complex with actin cytoskeleton Phosphorylated ITAMs Recruit ZAP70 Permitting Lck to Phosphorylate and Activate ZAP70 ZAP-70 Phosphorylates Scaffold Proteins LAT and SLP-76 to Activate Phospholipase C- Phospholipase C- Activates Protein Kinase C and Ras-GEF (RasGRP) ZAP-70 Initiates Cascade Activating Guanine-nucleotide Exchange (GEF) Factors That Activate Ras Transduction of the Signal Into the Nucleus Activates Genes There are multiple pathways that transduce the signal from the cytoplasm into the nucleus Different transcription factors activate different groups of genes MAP Kinase Cascade Transduces a Signal Into the Nucleus Ras activates MAP kinase kinase kinase (Raf). Raf phosphorylates serine/threonine on MAP kinase kinase (Mek) and activates it. MAP kinase kinase (Mek) phosphorylates threonine and tyrosine on MAP kinase (Erk) thereby activating it. MAP kinase (Erk) activates the Elk transcription factor which stimulates Fos gene transcription. Calcium Induces the Translocation of the Transcription Factor NFAT Into the Nucleus Phosphorylation of IB Releases the Transcription Factor NFB Thereby Enabling It to Translocate Into the Nucleus T Cell and B Cell Activation Requires Two Signals Co-stimulatory Signal Activates a Parallel MAPK Path That Activates Complementary Genes Ras activates MAP kinase kinase kinase (Raf). Raf phosphorylates serine/threonine on MAP kinase kinase (Mek) and activates it. MAP kinase kinase (Mek) phosphorylates threonine and tyrosine on MAP kinase (Erk) thereby activating it. MAP kinase (Erk) activates the Elk transcription factor which stimulates Fos gene transcription. Stimulatory Signal Pathways Integrated by Production of Fos and Jun- Components of the Heterodimer AP-1 Transcription Factor Broad Range of Nuclear Factors Are Activated in TCR Signal Transduction Cascade Initiation of TCR-mediated signals Biochemical intermediates Active enzymes Transcription factors Abbas- Cellular and Molecular Immunology Activation of Transcription Factors To Turn on Genes by Activation of Kinases and Phosphatases Abbas- Cellular and Molecular Immunology Cyclosporin and Tacrolimus Block Calcineurin Signaling and Thereby Suppress T Cell Activation In 1984, Fujisawa scientists discovered tacrolimus in a soil sample taken from Mt. Tsukuba in Japan. Not long after, scientists realized that some of the properties of tacrolimus may help treat certain conditions and diseases. Eventually, they saw that this drug could be used to safely treat patients with moderate to severe eczema—thus, pioneering the use of topical immunomodulators (TIMs). Protopic was the first of this new class of nonsteroid drugs approved for the treatment of moderate to severe eczema—giving doctors the first prescription alternative for the treatment of moderate to severe eczema in 40 years. Cross-linking Surface Immunoglobulin Activates B Cells Cross-linking Surface Immunoglobulin Activates ITAMs Questions to Consider After recognition of its cognate MHC:peptide, how does the T cell receptor activate immune response genes? What are the structural motifs used by signal transduction molecules that permit specific interaction and activation of effector proteins? How do genes know what is going on in the outside environment requiring them to make their proteins at the appropriate time?