* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download document 8916677
Survey
Document related concepts
Transcript
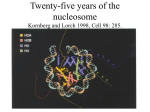
Overarching goal of the lab: • Understand how identical genotypes in identical environments give rise to different phenotypes The organization of the human genome in the nucleus is almost unbelievable Image by Christian Bouvier The functional organization of the human genome Jonathan Dennis, FSU Biological Science FSU College of Medicine, Grand Rounds 21 April 2011 Agenda • Introduction to chromatin structure • Techniques to assay chromatin structure • RESULTS – The molecular pharmacology of the anti‐ inflammatory butyrate – Chromatin structural biomarkers of cancer tumor grade Agenda • Introduction to chromatin structure • Techniques to assay chromatin structure • RESULTS – The molecular pharmacology of the anti‐ inflammatory butyrate – Chromatin structural biomarkers of cancer tumor grade We know surprisingly little about the organization of the human genome Image by Christian Bouvier The organization of the human genome must occur at multiple levels Analogy to levels of protein structure The organization of the human genome must occur at multiple levels Analogy to levels of protein structure We know surprisingly little about the organization of the human genome PRIMARY STRUCTURE Image by Christian Bouvier We know the structure of the core particle Crystal structure of the nucleosome core particle at 2.8 Å resolution Karolin Luger, Armin W. Mäder, Robin K. Richmond, David F. Sargent and Timothy J. Richmond Nature 389, 251‐260(18 September 1997) The primary structure of chromatin is regulated multiple ways Local chromatin architecture – How are nucleosomes distributed to give appropriate access to DNA sequence? Two examples: I. ATP‐dependent remodelers II. Covalent modification of histones The primary structure of chromatin is regulated multiple ways Local chromatin architecture – How are nucleosomes distributed to give appropriate access to DNA sequence? Two examples: I. ATP‐dependent remodelers II. Covalent modification of histones The primary structure of chromatin is regulated multiple ways I. ATP‐dependent remodelers ATP-dep rem. The primary structure of chromatin is regulated multiple ways I. ATP‐dependent remodelers ATP ADP ATP-dep rem. The primary structure of chromatin is regulated multiple ways I. ATP‐dependent remodelers ATP dependent remodelers regulate the expression of several genes Local chromatin architecture – How are nucleosomes distributed to give appropriate access to DNA sequence? Ordered Recruitment of Chromatin Modifying and General Transcription Factors to the IFN-β Promoter Agalioti, Lomvardas, Parekh,Ye, Maniatis, and Thanos Cell, Volume 103, Issue 4, 10 November 2000, Pages 667-678 The primary structure of chromatin is regulated multiple ways Local chromatin architecture – How are nucleosomes distributed to give appropriate access to DNA sequence? Two examples: I. ATP‐dependent remodelers II. Covalent modification of histones Histones may be covalently modified by protein complexes II. Covalent modification of histones Histones may be covalently modified by protein complexes II. Covalent modification of histones Histones may be covalently modified by protein complexes II. Covalent modification of histones Histones may be covalently modified by protein complexes II. Covalent modification of histones •acetylation •methylation •ubiquitylation •and many more… Histones may be covalently modified by protein complexes II. Covalent modification of histones •acetylation H3 K36: transcription •methylation H3 K4: gene activation •ubiquitylation H2A K119: Hox cellular memory •and many more… and many combinations We know surprisingly little about the organization of the human genome SECONDARY STRUCTURE Image by Christian Bouvier Models of the chromatin fiber are controversial Robinson P J J et al. PNAS 2006;103:6506-6511 ©2006 by National Academy of Sciences We know surprisingly little about the organization of the human genome TERTIARY STRUCTURE Image by Christian Bouvier Very little is known about the tertiary structure of chromosomes Global chromatin architecture – How are chromosomes organized to give appropriate access to soluble regulatory factors? LESS ACCESSIBLE MORE ACCESSIBLE Very little is known about the tertiary structure of chromosomes Global chromatin architecture – How are chromosomes organized to give appropriate access to soluble regulatory factors? CELL TYPE A CELL TYPE B The organization of the human genome must occur at multiple levels Global chromatin architecture – How are chromosomes organized to give appropriate access to soluble regulatory factors? We know surprisingly little about the organization of the human genome QUATERNARY STRUCTURE Image by Christian Bouvier Chromosomes occupy fixed spaces in the human nucleus Global chromatin architecture – How are chromosomes organized to interact with one another Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, et al. 2005 Three-Dimensional Maps of All Chromosomes in Human Male Fibroblast Nuclei and Prometaphase Rosettes. PLoS Biol 3(5): e157. A system for the simultaneous assessment of local and global chromatin architecture MOTIVATIONS: •What is the relationship between chromatin structure (both at the local and global level) and regulation of nuclear events? •To what degree are local changes in nucleosome distribution, linked with global changes in chromatin accessibility? A system for the simultaneous assessment of local and global chromatin architecture REQUIREMENTS: •High‐throughput – Don’t just look at one cell type. Look at dozens of cell types •Cost‐effective – The price must not be prohibitive •Robust – The signal must be reliably interpretable A system for the simultaneous assessment of local and global chromatin architecture REQUIREMENTS: •High‐throughput – Don’t just look at one cell type. Look at dozens of cell types (12 samples simultaneously) •Cost‐effective – The price must not be prohibitive (~$200‐$300 per sample) •Robust – The signal must be reliably interpretable (consistent with previous findings and between experiments) Agenda • Introduction to chromatin structure • Techniques to assay chromatin structure • RESULTS – The molecular pharmacology of the anti‐ inflammatory butyrate – Chromatin structural biomarkers of cancer tumor grade Techniques to assay chromatin structure We use a single nuclease (MNase) to assay • Primary structure – nucleosome distribution • Tertiary structure – chromosome accessibility Techniques to assay chromatin structure We use a single nuclease (MNase) to assay • Primary structure – nucleosome distribution • Tertiary structure – chromosome accessibility Nuclease as a probe of nucleosome distribution– Micrococcal nuclease is an internucleosomal cutter Nuclease as a probe of nucleosome distribution – Micrococcal nuclease is an internucleosomal cutter Nuclease as a probe of nucleosome distribution – Micrococcal nuclease is an internucleosomal cutter [MNase] 4.3 kbp 2.0 kbp 7N 6N 5N 4N 3N 500 bp 2N N Brian Spetman Nuclease as a probe of nucleosome distribution – Micrococcal nuclease is an internucleosomal cutter [MNase] 4.3 kbp 2.0 kbp 7N 6N 5N 4N 3N 500 bp 2N N Brian Spetman Nuclease as a probe of nucleosome distribution – Micrococcal nuclease is an internucleosomal cutter [MNase] 4.3 kbp 2.0 kbp 7N 6N 5N 4N 3N 500 bp 2N N Brian Spetman Nuclease as a probe of nucleosome distribution – nucleosome distribution determined by microarray [MNase] 4.3 kbp 2.0 kbp 7N 6N 5N 4N 3N 500 bp 2N N Each “subarray” contains 135,000 probes that cover up to 500 TSS at 13bp resolution ~150 ~ bp DNA nucleosome posi oning nucleosome posi oning Nuclease as a probe of nucleosome distribution – readout = log (nucleosomal/bare genomic) TSS ‐1 NFR +1 genomic posi on TSS ‐1 NFR +1 genomic posi on nucleosome posi oning nucleosome posi oning Nuclease as a probe of nucleosome distribution – readout = log (nucleosomal/bare genomic) TSS ‐1 NFR +1 genomic posi on IDEALIZED TSS ‐1 NFR +1 genomic posi on CD69 TSS U937 (human histiocytic lymphoma) Brian Spetman Nuclease as a probe of nucleosome distribution – We can observe TSS poising or activation Unstimulated U937 Imiquimod (viral) stimulated Allows us to characterize the promoter architecture associated with specific classes of genes Brittany Sexton Techniques to assay chromatin structure We use a single nuclease (MNase) to assay • Primary structure – nucleosome distribution • Tertiary structure – chromosome accessibility Nuclease as a probe of chromatin accessibility Nuclease as a probe of chromatin accessibility – accessible areas are more readily cut Nuclease as a probe of chromatin accessibility – accessible areas are separable by electrophoresis Nuclease as a probe of chromatin accessibility – genomic coordinates of [in]accessible determined by microarray LESS ACCESSIBLE Label with Cy5 MORE ACCESSIBLE Label with Cy3 Nuclease as a probe of chromatin accessibility – genomic coordinates of [in]accessible determined by microarray LESS ACCESSIBLE Label with Cy5 MORE ACCESSIBLE Label with Cy3 Nuclease as a probe of chromatin accessibility – genomic coordinates of [in]accessible determined by microarray LESS ACCESSIBLE Label with Cy5 MORE ACCESSIBLE Label with Cy3 Each “subarray” contains 135,000 probes that cover the entire human genome at 12.5 kb resolution Nuclease as a probe of chromatin accessibility ‐ readout = log (inaccessible/accessible) log(inaccessible/accessible) Whole human genome Chr. less accessible more accessible 1 2 3 4 5 6 7 8 9 10 12 14 16 20 Jurkat ‐ T lymphocyte cells Crystal Pickeral Nuclease as a probe of chromatin accessibility ‐ readout = log (inaccessible/accessible) log(inaccessible/accessible) Whole human genome Chr. less accessible more accessible 1 2 3 4 5 6 7 8 9 10 12 14 16 20 Jurkat ‐ T lymphocyte cells Crystal Pickeral Nuclease as a probe of chromatin accessibility ‐ readout = log (inaccessible/accessible) log(inaccessible/accessible) Chromosome 1 less accessible more accessible Jurkat ‐ T lymphocyte cells Crystal Pickeral CELLS EXPRESSION ACCESSIBILITY NUCLEOSOME Agenda • Introduction to chromatin structure • Techniques to assay chromatin structure • RESULTS – The molecular pharmacology of the anti‐ inflammatory butyrate – Chromatin structural biomarkers of cancer tumor grade Histone Deacetylase Inhibitors Prevent Nucleosome Redistributions at Transcription Start Sites Crystal Pickeral • FSU Undergraduate Senior (for about 10 more days) • FSU College of Medicine Student (in about six weeks) Inflammatory Bowel disease is a prevalent autoimmune disorder with an unknown cause genetic immunological IBD dysbiosis infectious Chronic inflammation associated with IBD is caused by dysregulation of the mucosal immune system • Mucosal immune system – Depends on delicate balance of commensal bacteria and harmful pathogens – IBD patients have irregularities in their microflora Frank DN, St. Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. (2007) Molecular‐phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci 34:13780‐13785 One of the important roles of commensal bacteria is aiding in digestion • Breaking down of complex carbohydrates into short chain fatty acids • Butyrate is a short chain fatty acid that provides energy to intestinal epithelial cells • Members of Firmicutes phylum are the main butyrate producing bacteria Frank DN, St. Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. (2007) Molecular‐phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci 34:13780‐13785 Butyrate is a powerful histone deacetylase inhibitor (HDACi) • Histone tails can be modified by acetylation, methylation and phosphorylation • These modifications affect the interactions between DNA and the histones that ultimately affect the organization of chromatin. • Butyrate is a powerful histone deacetylase inhibitor Sparmann A, Lohuizen MV. 2006. Polycomb silencers control cell fate, development and cancer. Nature Reviews Cancer 6:846‐856 Butyrate blocks the removal of acetyl groups from histone tails leading to a hyperacetylated state. Johnstone RW. 2002. Histon‐deacetylase inhibitors: novel drugs for the treatment of cancer. Nature Reviews:Drug Discovery 1:287‐299 Butyrate provides a connection between chromatin structure and dysbiosis Gut lumen “good” bacteria “bad” bacteria Carbohydrates butyrate Epithelial Cells butyrate Macrophage HDACi AC AC AC HDACi are classified according to structure Hydroxamate‐based HDACi Kazantsev AG, Thompson LM. 2008. Therapuetic application of histone deacetylase inhibitors for central nervous system disorders. Nature Reviews Drug Discovery 7:854‐868 Sodium butyrate Valproic acid (VPA) • Hydroxamate‐based class 1 HDAC inhibitor • Hydroxamate‐based class 1 HDAC inhibitor • Butyrate is produced in the gut by commensal bacteria. • Used to treat certain types of seizures, mania in people with bipolar disorder and to prevent migraine headaches. • Butyrates are common treatments for Inflammatory Bowel Disease (IBD) • Being tested in treatments for inflammatory diseases. HDACi have anti‐inflammatory properties • Butyrate decreases the production of TNF‐ and IL‐12 • Valproic acid decreases the production of INF‐ in a concentration dependent manner Saemann MD, Bohmig GB, et. Al.2000. Anti‐inflammatory effects of sodium butyrate on human monocytes: potent inhibition of IL‐12 and up‐regulation of IL‐10 production. The FASEB journal 14:2380‐2382 HDACi have potent anti‐inflammatory properties • The mechanism behind these anti‐inflammatory properties is unknown Predict that butyrate and VPA could operate via similar mechanisms • Compounds with similar structures VPA: Butyrate: • Both act by blocking the machinery that would remove acetyl groups from histones • But are administered for very different reasons • Investigating the effect of these drugs on chromatin structure will provide insight into the molecular pharmacology of these drugs. Experimental goal • The goal of this experiment is to understand the relationship between the anti‐ inflammatory properties of histone deacetylase inhibitors and the changes they induce in chromatin structure in a resting state and during an immune insult. Significance • We propose that the study of the effect of class 1 HDAC inhibitor treatment on chromatin structure will illuminate the pathology of inflammatory disorders as well as provide insight into the mechanisms, behind how HDAC inhibitors are effective treatments. Treatment and harvesting of cells PMA differentiated U937 cells 1x107/experiment Time (hours) Untreated +1mM VPA +2mM NAB +LPS 4 hours 8 hours +LPS +LPS 4 hours Untreated NAB NAB+LPS VPA+LPS VPA LPS LPS is a strong inducer of the inflammatory response • Stimulating cells with LPS will allow us to challenge the anti‐ inflammatory effects of HDACi during an immune response LPS Pro‐inflammatory Response LPS is a strong inducer of the inflammatory response • Stimulating cells with LPS will allow us to challenge the anti‐ inflammatory effects of HDACi during an immune response LPS MAP Kinase NF‐kB TNF‐ IL‐6 ICAM‐1 Pro‐inflammatory Response Experimental design UN LPS B BL V VL Statistical analysis allows objective determination of changes in nucleosomal distributions 0.0 -1.0 -0.5 Signal 0.5 1.0 1.5 IL6 22765700 22766000 22766300 22766600 22766900 22767200 22767500 22767800 1.0 0.0 ‐ -1.0 + 0 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 0 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 0.0 1.0 2.0 t-test Correlation Position HDAC Inhibitors have limited effects on nucleosome redistributions at TSS HDACi Number of genes with nuc. Redistributions Percent of genes with nuc. redistributions Butyrate 23/505 4.55% Valproic acid 22/505 4.3% IL8 Log2 (mononucleosomal/bare genomic) CCL2 Log2 (mononucleosomal/bare genomic) HDAC Inhibitors induce modest changes in nucleosome distribution Genomic position (bp) Genomic position (bp) Nucleosomal redistributions that occur show HDACi specificity NaB (23) 17 VPA (22) NaB + VPA 6 16 Nucleosomal redistributions that occur show HDACi specificity 3 TMPRSS11D butyrate valproic acid butyrate valproic acid -1.0 -1 -0.5 -0.5 0 0.0 0.0 1 0.5 0.5 2 1.0 1.0 1.5 butyrate valproic acid IL8 2 5 -1.0 Log2 (mononucleosomal/bare genomic) CCL2 Genomic position (bp) HDACi’s decrease pro‐inflammatory protein expression Cytokine produc on 80 70 pg/ml 60 50 40 IL6 30 TNF‐alpha 20 10 0 Un butyrate VPA Figure 6. Concentra ons of IL6 and TNF‐alpha secreted in media from untreated, 2mM butyrate, and 1mM valproic acid treatments as determined by ELISA. LPS serves as a reliable immune insult model • Component of bacterial cell walls • Strong inducer of the inflammatory response • Stimulating cells with LPS will allow us to challenge the anti‐ inflammatory effects of HDACi during an immune response LPS MAP Kinase NF‐kB TNF‐ IL‐6 ICAM‐1 Pro‐inflammatory Response HDACi’s limit the number of nucleosome redistributions during an immune insult Stimulus Number of genes with nuc. Redistributions Percent of genes with nuc. redistributions LPS 455/505 90.1% Butyrate +LPS 65/505 12.8% Valproic acid +LPS 70/505 13.8% HDACi’s block nucleosome redistributions induced by an immune insult TREM1 1.0 0.5 0.0 -0.5 -1.0 -1.5 Log2(monoucleosomal/bare genomic) 1.5 Butyrate Triggers release of pro‐inflammatory 500 TSS +500 chemokines and cytokines. Genomic position (bp) ‐ TREM1 1.0 0.5 0.0 -0.5 -1.0 1.5 Log2(monoucleosomal/bare genomic) 1.5 Butyrate LPS Triggers release of pro‐inflammatory 500 TSS +500 chemokines and cytokines. Genomic position (bp) ‐ TREM1 1.0 0.5 0.0 -0.5 -1.0 -1.5 Log2(monoucleosomal/bare genomic) 1.5 Butyrate Butyrate+LPS Triggers release of pro‐inflammatory 500 TSS +500 chemokines and cytokines. Genomic position (bp) ‐ IL10 1.0 0.5 0.0 -0.5 -1.0 -1.5 Log2(monoucleosomal/bare genomic) 1.5 Butyrate LPS 500 TSS +500 This cytokine has pleiotropic effects in Genomic position (bp) immunoregulation and inflammation. ‐ IL10 1.0 0.5 0.0 -0.5 -1.0 -1.5 Log2(monoucleosomal/bare genomic) 1.5 Butyrate Butyrate+LPS 500 TSS +500 This cytokine has pleiotropic effects in Genomic position (bp) immunoregulation and inflammation. ‐ CX3CL1 1.0 0.5 0.0 -0.5 -1.0 -1.5 -2.0 Log2(monoucleosomal/bare genomic) 1.5 Butyrate LPS Chemotactic for T‐cells and monocytes. ‐ 500 TSS +500 Genomic position (bp) CX3CL1 1.0 0.5 0.0 -0.5 -1.0 -1.5 -2.0 Log2(monoucleosomal/bare genomic) 1.5 Butyrate Butyrate+LPS Chemotactic for T‐cells and monocytes. ‐ 500 TSS +500 Genomic position (bp) IL2RA 1.0 0.5 0.0 -0.5 -1.0 -1.5 -2.0 Log2(monoucleosomal/bare genomic) 1.5 Butyrate LPS Receptor for IL2 which is required for T‐ cell proliferation and other activities crucial to regulation of the immune response. ‐ 500 TSS +500 Genomic position (bp) IL2RA 1.0 0.5 0.0 -0.5 -1.0 -1.5 -2.0 Log2(monoucleosomal/bare genomic) 1.5 Butyrate Butyrate+LPS Receptor for IL2 which is required for T‐ cell proliferation and other activities crucial to regulation of the immune response. ‐ 500 TSS +500 Genomic position (bp) -0.5 0.0 0.5 1.0 1.5 Butyrate LPS -1.0 Log2(monoucleosomal/bare genomic) IL6 ‐ A cytokine that functions in inflammation. 500 TSS +500 Genomic position (bp) -0.5 0.0 0.5 1.0 1.5 Butyrate Butyrate+LPS -1.0 Log2(monoucleosomal/bare genomic) IL6 A cytokine that functions in inflammation. ‐ 500 TSS +500 Genomic position (bp) HDACi’s have cytokine specific effects Cytokine produc on 1800 1600 1400 pg.ml 1200 1000 IL6 800 TNF‐alpha 600 400 200 0 LPS butyrate +LPS VPA +LPS Figure 8. Concentra ons of IL6 and TNF‐alpha secreted in media from LPS, butyrate + LPS, and valproic acid + LPS treatments, as determined by ELISA. HDACi “lock‐in” chromatin structural states at TSS • 90% of genes had nucleosomal redistributions when stimulated with LPS. • 13% of genes had nucleosomal redistributions with HDACi‐LPS co‐treatments. • 70% of the loci showing LPS‐induced nucleosome redistributions were inhibited by HDACi pretreatment. IL‐6 displays a canonical promoter architecture upon activation nucleosome posi oning nucleosome posi oning LPS treated TSS ‐1 NFR +1 +2 genomic posi on IDEALIZED +3 ‐1 NFR +1 +2 +3 genomic posi on IL‐6 TSS Figure 15. An idealized promoter structure shown in green consisting of a nucleosome free region (NFR) flanked by the ‐1 nucleosome upstream of the TSS and the +1 nucleosome downstream of the TSS. The promoter structure of activated IL‐6 is shown in red. This canonical promoter structure is associated with activation pg/ml 800 un 600 LPS 400 200 0 un 1.5 1.0 0.5 0.0 1000 -0.5 1200 Untreated LPS -1.0 IL‐6 Protein Expression Log2(mononucleosomal/ bare genomic) IL6 LPS Genomic Position Reduction in IL‐6 protein expression is linked to changes in chromatin structure LPS 600 Butyrate +LPS 400 200 0 LPS Butyrate +LPS 1.5 1.0 0.5 pg/ml 800 0.0 1000 -0.5 1200 LPS Butyrate +LPS -1.0 IL‐6 Protein Expression Log2(mononucleosomal/ bare genomic) IL6 Genomic Position HDACi’s block LPS‐induced changes pg/ml un 30 Butyrate +LPS 20 10 0 un Butyrate +LPS 1.5 1.0 0.5 40 0.0 50 -0.5 60 Untreated Butyrate +LPS -1.0 IL‐6 Protein Expression Log2(mononucleosomal/ bare genomic) IL6 Genomic Position IL‐6 is a key pro‐inflammatory cytokine • Activates acute phase proteins, which are key molecules in regulating the immune response • Serve as effective markers of severity of disease state • High expression of IL‐6 is seen in rheumatoid arthritis and IBD patients HDACi’s block nucleosome redistributions induced by an immune insult This is the first report linking the anti inflammatory properties of HDAC inhibitors to the alterations in the chromatin regulatory structural potentials. HDACi’s block nucleosome redistributions induced by an immune insult Further studies of this nature will help to illuminate the pathology of immune diseases and guide the identification of effective and targeted treatments. Agenda • Introduction to chromatin structure • Techniques to assay chromatin structure • RESULTS – The molecular pharmacology of the anti‐ inflammatory butyrate – Chromatin structural biomarkers of cancer tumor grade Chromatin Structural Changes Serve as Effective Biomarkers for Cancer Grade Brooke Roberts Druliner • FSU Biological Science Graduate Student Tumor grades signify the differentiation status of tumor cells GRADE DESCRIPTION G1 low Well‐differentiated G2 intermediate Moderately differentiated G3 high Poorly differentiated G4 high Undifferentiated We obtained the following lung adenocarcinoma tumor samples with corresponding normal tissue Sample ID Date Organ Cancer Type Grade Age Gender 0620T 1/10/07 Lung Adenocarcinoma 1 55 Male 1357T 2/28/08 Lung Adenocarcinoma 1 63 Female 1294T 1/10/08 Lung Adenocarcinoma 2 67 Female 2123T 8/20/09 Lung Adenocarcinoma 2 58 Female 0873T 5/24/07 Lung Adenocarcinoma 3 51 Male 1831T 3/5/09 Lung Adenocarcinoma 3 75 Male TISSUE EXPRESSION ACCESSIBILITY NUCLEOSOME Low grade tumor samples show many changes in nucleosome distribution TSS out of 415 showing changes in nucleosome distribution (tumor compared to normal) GRADE 1 GRADE 2 GRADE 3 400 350 337 300 250 200 150 180 100 61 50 35 0 0620_G1 1357_G1 1294_G2 26 2123_G2 0873_G3 Low grade tumor samples show many changes in nucleosome distribution GRADE 1 GRADE 3 2 1 −1 −2 108092900 108093300 Black line= Normal Red line = Tumor −3 108092500 0 Signal 0 −3 −2 −1 Signal 1 2 3 chr11 3 chr11 108093700 108092500 Position ATM 108094100 108094500 Black line= Normal Red line = Tumor 108092900 108093300 108093700 108094100 108094500 Position ATM Ataxia telangiectasia mutated (ATM) gene. The protein made by the ATM gene functions to control the rate at which cells grow. Low grade tumor samples show few changes in nuclease accessibility GRADE 1 Chrom. 1 2 3 4 5 6 7 8 9 10 11 12 13 14 16 18 20 2 3 4 5 6 7 8 9 10 11 12 13 14 16 18 20 GRADE 1 Chrom. 1 Intermediate grade tumor samples show few changes in nuclease accessibility GRADE 2 Chrom. 1 2 3 4 5 6 7 8 9 10 11 12 13 14 16 18 20 2 3 4 5 6 7 8 9 10 11 12 13 14 16 18 20 GRADE 2 Chrom. 1 High grade tumor sample shows major changes in nuclease accessibility GRADE 3 Chrom. 1 2 3 4 5 6 7 8 9 10 11 12 13 14 16 18 20 High grade tumor sample shows major changes in nuclease accessibility GRADE 3 Chrom. 1 2 3 4 5 6 7 8 9 10 11 12 13 14 16 18 20 2 3 4 5 6 7 8 9 10 11 12 13 14 16 18 20 GRADE 1 Chrom. 1 An initial model for chromatin structural biomarkers for tumor severity GRADE 1 2 3 Nucleosome distribution Chromosome accessibility gene expression cell type Perhaps chromatin structural biomarkers may serve as indicators of cancer grade • Independent of genotype • Independent of gene expression • Ability to differentiate specific grade subtypes Thank you to the individuals who did the work and developed these techniques Ph.D. Students Brooke R. Druliner Justin Fincher (CS) Sarah Lueking Brian Spetman Undergraduates Ely Gracia Preston Hood Josh Koerner Tarreq Noori Crystal Pickeral Brittany Sexton Postdoctoral Fellow Parwez Alam Immunology Collaborator Marie Charrel Dennis Funding FSU Department of Biological Science FSU CRC FYAP Program Questions? Thank you It will be interesting to understand the relationships between regulation at the gene locus level and regulation at the whole genome level that occurs upon HDACi treatment 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 x Untreated VPA Log2(Inacessible/accessible) Untreated butyrate 0.0e+00 8.0e+08 1.2e+09 1.6e+09 2.0e+09 2.4e+09 0.0e+00 2.8e+09 1.6e+09 2.0e+09 2.4e+09 2.8e+09 Untreated VPA + LPS 4.0e+08 8.0e+08 1.2e+09 1.6e+09 2.0e+09 2.4e+09 2.8e+09 Log2(Inacessible/accessible) 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 x ‐4 ‐2 0 2 Log2(Inacessible/accessible) C) 4.0e+08 1.2e+09 Log2(Inacessible/accessible) Untreated Butyrate + LPS 0.0e+00 8.0e+08 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 x 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 x ‐2 0 2 Log2(Inacessible/accessible) B) 4.0e+08 Untreated LPS Untreated LPS 0.0e+00 4.0e+08 8.0e+08 1.2e+09 1.6e+09 Genomic posi on 2.0e+09 2.4e+09 2.8e+09 0.0e+00 4.0e+08 8.0e+08 1.2e+09 1.6e+09 Genomic posi on 2.0e+09 2.4e+09 2.8e+09