* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download 7. Sporulation
Survey
Document related concepts
Transcript
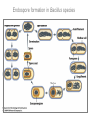
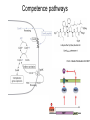
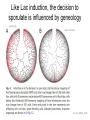
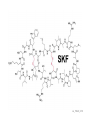
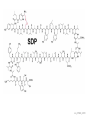
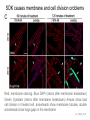
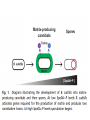
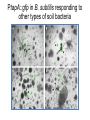
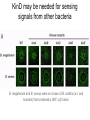
Endospore formation in Bacillus species Diagram of sporulation Genes involved in sporulation Phosphotransfer during sporulation Phosphotransfer during initiation of sporulation Note the inhibition of an inhibitor (Spo0A-P inhibits ArbB) -this leads to an increase in antibiotics, biofilm formation etc. Regulation of sporulation by dephosphorylation Effects of PhrA peptides on sporulation Competence pathways From: Okada Tetrahedron 62:8907 Competence pathways (more complex view) http://www.mun.ca/biochem/courses/41 03/topics/transformation.html. This site has a good summary of what all of these proteins do. ComA-P induces rapA transcription-this means that sporulation is inhibited when the competence state is predominant Fig. 1. Schematic representation of the regulatory network that governs sporulation initiation in Bacillus subtilis. Phosphate input into the phosphorelay signal transduction system is provided by the two histidine kinases, KinA and KinB. Activation of KinB requires the KapB lipoprotein[43]. KinA is negatively regulated by the Kipl inhibitor, whose activity is counteracted by the KipA anti-inhibitor[18]. Accumulation of the phosphorylated Spo0F (Spo0FP) intermediate response regulator is modulated by the RapA and RapB phosphatases. RapA expression is induced by the ComA response regulator for competence development ,while its activity is modulated by the PhrA peptide. An exportミimport control circuit regulates the production of the PhrA pentapeptide inhibitor, which, upon reimportation by the Opp transport system, directly and specifically inhibits RapA phosphatase activity. The Spo0E phosphatase is the final checkpoint and modulates the level of Spo0AP by direct dephosphorylation. Spo0AP is both an activator of sporulation genes and a repressor of genes that prevent sporulation. From: Perego Trends Micro 9:366 Behavior of B. subtilis during colony growth Fig. 1. Typical B. subtilis microcolony development. (A) Still frames (phase contrast) of the outgrowth of a single cell into a sporulating microcolony. (Insets) Magnifications. (B) Log of microcolony biomass (black line) and average growth rates (gray triangles) are plotted in time. The growth rate is calculated as an exponential fit to cell length in time (arbitrary units, AU), measured from the birth point of the cell until the next cell division event or cell fate decision (see SI Materials and Methods). Biomass was calculated as a function of cell length (AU). (C) Each circle represents the point of birth in time of an individual cell. The average growth rate of this cell during its life is represented on the y axis (AU) Behavior of B. subtilis during colony growth Cell fates plotted onto the birth points of C. Every birth point shown indicates the birth of a cell for which it is certain that either this cell or all of its descendents will follow a specific fate. Blue circles show the growth rates of individual cells committed to spore formation; green triangles show diauxic growth fate cells; and red triangles indicate lysing cells. B. subtilis grows more slowly as it ages Cells of different fates express sporulation gene spoIIA to different degrees Fluorescence of a spoIIA::gfp fusion. Like Lac induction, the decision to sporulate is influenced by geneology spoIIA::gfp spore formation Veening_PNAS_2008 A: An IPTG inducible promoter is used to drive the spoOA gene and give normal amounts of this key regulator. Colonies show a variety of cell types (See panel C) B: The spo0A mutant sad67 is always active (like it was phosphorylated). It does not give a variety of cell types (see panel D) Thus proper control of the phosphorylation state is important for the generation of different subtypes of B. subtilis (growers, lysers and sporulators. Overproducing RapA is similar to the sad67 phenotype—Why? Imaging MS of SDK and SKF Fig. 1. IMS of intraspecies metabolic exchange. (A) IMS of PY79 and Δspo0A (KP648) coculture. (1) Ion distributions observed for surfactin ([M+K]+), SKF ([M+H]+), and SDP ([M+K]+). 1i is a superimposition of all three ions. (2) Superimposition of the photograph and IMS data. Arrows point to glassy region of Δspo0AandSDP overlap.1 Liu_PNAS_2010 Liu_PNAS_2010 Liu_PNAS_2010 SDK added to growing cultures of B. subtilis 0, 0.2, 2 ug/ml 5 ug/ml 10 ug/ml SDK added at various concentrations at T=0 to Dspo0A 20 ug/ml SDK added at 20 ug/ml at various times to Dspo0A Liu_PNAS_2010 SDK causes membrane and cell division problems Red: membrane staining. Blue DAPI (stains after membrane breakdown) Green Cytostain (stains after membrane breakdown) Arrows show bad cell division in treated cell, arrowheads show membrane tubules, double arrowheads show large gaps in the membrane Liu_PNAS_2010 SDP causes cell lysis/growth inhibition, SKF really doesn’t Fig. 4. Spot assays to compare the effect of exogenously supplied and endogenously produced SDP and SKF. Lawns of indicated strains were prepared in top agar. After solidification, the lawns were spotted with either (A) purified SDP, SKF, and DMSO or (B,C) the indicated strains that overexpress or lack SDP and SKF. Liu_PNAS_2010 SDK also kills Staphylococcus SDK at various concentration was added to cultures of various species. The final growth yields are plotted Surfactin and Nisin both activate EPS (biofilm) formation via KinC Surfactin: made by B. subtilis. It is a surfactant and an antibiotic Surfactin is a peptide, synthesized w/o ribosomes Nisin is also a cyclic peptide antibiotic, made by Lactobacteria species Schematic of sigma70 Domain 1 PtapA::gfp in B. subtilis responding to other types of soil bacteria B. subtilis made matrix in response mainly to other species of Bacillus Types and numbers of species that induced the PtapA::gfp reporter KinD may be needed for sensing signals from other bacteria B. megaterium and B. cereus were on a lawn of B. subtilis (w.t. and mutants) that contained a SKF::yfp fusion Two types of responses to other Bacillus species Type 1: Cells induce SKF in response to compounds secreted by other Bacillus species. This response requires KinD. Blue on next slide TypeII: Any cells that are not already making SKF are killed and only those making SKF are immune to SKF and related molecules coming from the other Bacillus species—these live and glow green. This does not require KinD and live. Orange on next slide Two types of responses to other Bacillus species Conclusion: Species close to B. subtilis make molecules similar to SKF, and SKF-making B. subtilis cells are immune and can grow. Species more distant from B. subtilis make toxic signaling compounds. These are sensed by B. subtilis KinD and are used to initiate defense, killing, matrix formation and sporulation via high levels of Spo0A-P