* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download gd T cells
Lymphopoiesis wikipedia , lookup
Cancer immunotherapy wikipedia , lookup
Polyclonal B cell response wikipedia , lookup
Major histocompatibility complex wikipedia , lookup
Molecular mimicry wikipedia , lookup
Innate immune system wikipedia , lookup
Adaptive immune system wikipedia , lookup
X-linked severe combined immunodeficiency wikipedia , lookup
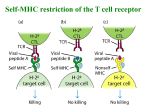
Chapter 9 T-Cell Receptor Interaction of ab TCR with class I MHC-peptide Dec 5, 2006 你需要學習的課題: 1. 2. 3. 4. T 細胞受體是如何發現的? 有什麼特性? ab TCR 與 gd TCR在辨認上有什麼不同? TCR 的基因及分子。 T 細胞辨識時,除了TCR 之外還有哪些 coreceptors ? T Cells Recognize Ag Only When Presented on the Membrane of a Cell by a Self-MHC Molecule This work was published in 1974. Zinkernagel and Doherty were awarded the Nobel prize In 1996. Identification of T-cell Receptors: 1. Clonotypic mAb how? 2. Gene cloning by subtractive hybridization Identification and Cloning of the TCR Genes ~ Hedrick and Davis, 1984 ~ Well-thought-out assumptions of TCR Genes : 1. mRNAs are associated with membrane-bound polyribosomes rather than with free cytoplasmic ribosomes. 2. mRNAs are only expressed in T, but not in B cells. subtractive hybridization (98% of the genes expressed in T and B cells are the same) 3. DNA is rearranged in mature T cells. Production and Identification of a cDNA Clone Encoding the TCR subtractive hybridization ~ 2% of total cDNA: 3% x 2% = 0.06% each of the 6 T-cell clones showed different Southern blot patterns. , and clones 3, 4, 5, 6, 7, 8, 9, 10 The cDNA clone 1 identified by the Southernblot analysis shown in the previous slide has all the hallmarks of a putative TCR gene: 1. It represents a gene sequence that rearranges. 2. It is expressed as a membrane-bound protein. 3. It is expressed only in T cells. The cDNA clone 1 is the b chain of the TCR. Later, cDNA clones were identified encoding theα chain, theγchain, and finally the δ chain. ab and gd T-cell Receptors: Structure and Roles Structural Similarity between mIgM and TCR Fab or gd T-cell receptor resembling an Fab fragment ? TCRab * TCRgd * The δ-chain gene segments are located between the Vα and Jα segments. a, g genes : V, J, C gene segments b, d genes : V, D, J, C gene segments Comparison of the gd TCR and ab TCR % of CD3+ T cells 1-10 % 90-99 % elbow angle → Contribute to differences in signaling mechanism and in how the molecules interact with coreceptors. (majority) gd T cells - In humans, the predominant receptor expressed on circulating gd cells recognizes a microbial phospholipid Ag, 3-formyl-1-butyl pyrophosphate, found on M. tuberculosis and other bacteria and parasites (similat to pattern recognition receptor?) - This specificity for frequently encountered pathogens led to speculation that gd cells may function as an arm of the innate immune response, allowing rapid reactivity to certain Ags without the need for a processing step. - The specificity of circulating gd cells in the mouse and of other species studied does not parallel that of humans, suggesting that the gd response may be directed against pathogens commonly encountered by a given species. - Since gd cells can secrete a spectrum of chemokines and cytokines, they may play a regulatory role in recruiting other cells to the site of invasion by pathogens. Ligands Recognized by gd T Cells: - gd T cells appear to bind directly to Ags without requiring Ag processing and presentation by MHC. - Some gd T cells may uniquely respond to heat-shock proteins and may have evolved to eliminate damaged cells as well as microbial invaders. Organization and Rearrangement of TCR Genes Germ-line Organization of the Mouse TCR a-, b-, g-, and d-chain Gene Segments d between Va and Ja : a productive rearrangement of a-chain gene segments deletes Cd Gene Rearrangements That Yield a Functional Gene Encoding the ab TCR The C region of TCR is much simpler than the C region of Ig genes: - TCR is expressed only in a membrane-bound form; thus, no differential RNA processing is required to produce membrane and secreted form. - TCRa has only a single C gene segment and TCRb has two C gene segments. - No known functional differences exist in C regions. - Although B cells and T cells use very similar mechanisms for V-region gene rearrangements, the Ig genes are not rearranged in T cells and the TCR genes are not rearranged in B cells. - The recombinase enzyme system is differently regulated in B and T cell lineage, so that only rearrangement of the correct receptor DNA occurs. - Chromatin is also uniquely re-configured in B cells and T cells to allow the recombinase access to Ig and TCR genes, respectively. Domains and CDRs of ab-TCR Comparison of Mechanisms for Generating Diversity in TCR Genes and Ig Genes The Location of One-turn (12-bp) and Two-turn (23-bp) Recognition Signal Sequences (RSS) in TCR b- and dchain DNA Differs from That in Ig H-chain DNA or V-D-D-D-J in humans generate considerable additional diversity in TCR genes. N-addition occurs in all the TCR genes. 1+41+42+43+44+45+46 =5461 - Although each junctional region in a TCR gene encodes only 10 to 20 a.a., enormous diversity can be generated in these regions. - The combined effects of P- and N- addition plus joining flexibility are estimated to be 1013 possible a.a. sequences in the TCR CDR3 region. abTCR: 3.0 x 103 x 4.6 x 102 = 1.4 x 106 T-cell Receptor Complex: TCR-CD3 T-Cell Receptor Complex: TCR-CD3 CD3 : ge + de + (90%) or or (10%) immunoreceptor tyrosine-based activation motif T-Cell Accessory Membrane Molecules Accessory Molecules Which Strengthen the Interaction between T Cells and APC (T cell) (APC) (costimulatory) CD4 and CD8 Coreceptors Bind to Conserved Regions of MHC Class II or I Molecules sometimes aa homodimer 55-kDa 30-38 kDa each chain class I CD8 CD4 class II Interaction of CD8 Coreceptor with TCR and Class I MHC Molecule a1 a2 Interaction of CD4 Coreceptor with TCR and Class II MHC Molecule Dissociation Constants (Kd) for Various Biological Systems (10-4 to 10-7) (10-6 to 10-10) Interactions between TCR/Peptide-MHC and Accessory Molecules/Ligands Two-point Contact - extracellular portion of CD4 : MHC intracellular CD4 : p56lck - Three-dimensional Structures of TCR-peptide-MHC Complexes Interaction between TCR and HLA-A2 with Bound HTLV-I Tax Peptide HTLV-1 tax peptide HLA-A2 MHC Molecule Viewed from Above HV loops of TCR-Vb Peptide HV loops of TCR-Va HLA-A2 Ternary Complex of TCR Bound to H-2Kb and Peptide TCR CDR1 & CDR2 of Va CDR3 of Va CDR1 & CDR2 of Vb CDR3 of Vb peptide H-2Kb Comparison of the Interactions between ab TCR and MHC-peptides 1. The angles at which the TCR molecule sits on the class I and class II MHC-peptide are different. 2. More number of contact residues between TCR and class II Alloreactivity of T Cells - a puzzling finding T cell recognition 1. self-MHC + foreign peptides + 2. allo-MHC + foreign peptides – self-MHC restriction Alloreactivity of T Cells - a puzzling finding T cell recognition 1. self-MHC + foreign peptides + 2. allo-MHC + foreign peptides – self-MHC restriction however, 3. allo-MHC ± allo-peptides +++ one explanation : cross-reactivity of 1 and 3 why? Models for Alloreactivity of T Cells