* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Document
Hardy–Weinberg principle wikipedia , lookup
Koinophilia wikipedia , lookup
Gene expression profiling wikipedia , lookup
Pharmacogenomics wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Gene expression programming wikipedia , lookup
Genetic testing wikipedia , lookup
History of genetic engineering wikipedia , lookup
Genetic engineering wikipedia , lookup
Medical genetics wikipedia , lookup
Genetic drift wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Public health genomics wikipedia , lookup
Designer baby wikipedia , lookup
Genome (book) wikipedia , lookup
Population genetics wikipedia , lookup
Microevolution wikipedia , lookup
Human genetic variation wikipedia , lookup
Behavioural genetics wikipedia , lookup
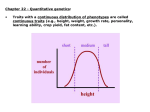
Chapter 18 Quantitative Genetics Continuous variation 13 - 18 April, 2005 Overview • In nature, variation in most phenotypic characters is continuous. • Such quantitative variation is studied with statistical techniques. • Continuous variation may be the result of segregation of interacting alleles at several loci with cumulative effect on phenotype. • Environmental interaction with genotype contributes to phenotypic variance. • Heritability is a population trait, not an individual one. Quantitative genetics •Genetics of continuously varying characters •Attempts to determine genetic variation contributing to character –number of loci with segregating alleles –how genes interact with one another and the environment (norm of reaction) –How do loci interact (dominance, epistasis, etc) –possible maternal effects •Often requires statistical analysis •Quantitative traits: small variation between genotypes, large variation within genotype. Statistical measures •Statistical distribution: description of set of quantitative measurements –graphical representation: e.g, histogram –distribution function: continuous curve •Mean: measure of central tendency (average) –mode: most frequent observation •Variance: measure of dispersion about mean •Correlation: relationship between two measured quantities Measures of central tendency Mode: most frequent class in sample bimodal: two frequent classes Mean: arithmetic average 1 Mean x x i N Measures of dispersion • Variance of sample: average squared deviation from mean 2 1 s ( xi x) N 1 2 • Standard deviation s variance s 2 Measures of relationship (1) • Correlation coefficient: relation between two variables, x and y – calculated from covariance of sample and standard deviations of x and y 1 cov xy ( xi x)( yi y ) N 1 cov xy correlatio n rxy sx s y R2=.82 R2=.99 Measures of relationship (2) • Correlation is estimate of precision of relation between two variables; can not be used to predict value of one given the other. • Regression describes relationship between two variables and allows their prediction. linear regression: y bx a Although most quantitative traits are polyfactorial, variation at a single locus can behave quantitatively. Norm of reaction • The relationship between environment and phenotype for a given genotype • Studied by subjecting homozygous lines to different environments – replicated (e.g., cloned) genotypes – inbred lines (e.g., from repeated sib mating) – use of dominant markers and crossover suppressors to make lines homozygous for particular chromosomes • Studies indicate that phenotypic differences between genotypes are small Heritability • Important to know whether genes contribute to phenotypic variation of quantitative character • Heritability is a population trait, not property of individual • Not same as familial trait shared by members of a family • Characters are heritable only if similarity arises from shared genotypes – estimated from phenotypic similarity of relatives – estimated by cosegregation of gene markers – difficult to estimate in humans 2 H: broad heritability s2p = total phenotypic variance in population s2g = genetic variance s2e = environmental variance cov = covariance between genotypic and environmental effects s2p = s2g + s2e + 2 cov ge H 2 s s 2 g 2 p s 2 g sg2 se2 H2 tells what part of population’s variation is attributable to genotypic variation. Estimating 2 H •Intercross homozygotes and measure variance within each heterozygous genotype. Average is s2e which can be subtracted from s2p to give s2g. •Statistically estimate from genetic similarities between relatives, particularly using difference in phenotypic correlation between monozygotic and dizygotic twins Meaning of 2 H • It is a population parameter, with no application to particular individuals • H2 > 0 means that genetic variance is present • H2 = 0 means that there is no genetic variation; genes may still be relevant to trait (as in development) • Value of H2 provides limited prediction about effect of environmental modification • H2 is specific to environmental conditions under which it is measured Locating genes •Difficult to identify genes for quantitative characters •Candidate gene: from prior biochemical or developmental knowledge •Quantitative trait loci (QTL) may be located to regions of chromosomes by cosegregation with marker genes •Dominance and epistasis may be involved 2 h: narrow heritability • Genetic variance can be divided into additive genetic variance and dominance variance 2 a 2 d s h 2 2 sa s se 2 • h2 is useful in determining whether there is selectable genetic variation – useful in plant and animal breeding Assignment: Concept map, Solved Problems 1-3, All Basic and Challenging Problems.