* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Proteases - Home - KSU Faculty Member websites
Point mutation wikipedia , lookup
Signal transduction wikipedia , lookup
Ultrasensitivity wikipedia , lookup
Peptide synthesis wikipedia , lookup
Western blot wikipedia , lookup
Oxidative phosphorylation wikipedia , lookup
Protein–protein interaction wikipedia , lookup
NADH:ubiquinone oxidoreductase (H+-translocating) wikipedia , lookup
Biochemistry wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Enzyme inhibitor wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Biosynthesis wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Metalloprotein wikipedia , lookup
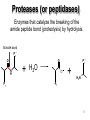
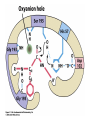
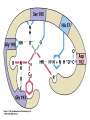
King Saud University College of Science Department of Biochemistry Disclaimer • The texts, tables and images contained in this course presentation are not my own, they can be found on: – References supplied – Atlases or – The web Mechanism of Enzyme Action BCH 321 Professor A. S. Alhomida King Saud University College of Science Department of Biochemistry Disclaimer • The texts, tables and images contained in this course presentation (BCH 320) are not my own, they can be found on: – References supplied – Atlases or – The web Mechanism of Enzyme Action Proteases Professor A. S. Alhomida 2 Proteases (or peptidases) Enzymes that catalyze the breaking of the amide peptide bond (proteolysis) by hydrolysis. Scissile bond P1' O O N H P1 + H2O P1' O + H3N + P1 3 Proteases are essential to physiologic processes • • • • • • • • • Inflammation Infection Fertilization Allergic reactions Cell growth and death Blood clotting Tumor growth Bone remodeling Etc… 4 Proteolytic enzymes are divided into two different categories • Limited proteolysis: a protease cleaves only one or a limited number of peptide bonds of a target protein leading to the activation or maturation of the formerly inactive protein. – e.g. conversion of prohormones to hormones. • Unlimited proteolysis: in which proteins are degraded into their amino acid constituents. – Ubiquitin/proteasome pathway: First conjugated to multiple molecule of the polypeptide ubiquitin. This modification marks them for rapid hydrolysis by the proteasome in the presence of ATP. – Lysosome pathway: Proteins are transferred into lysosomes which contain proteases that completely digests the protein into the amino acids. • These unlimited proteolysis pathways are essential for protein quality control and the reuse of amino acids during starvation. 5 Some Cells Secrete proteases • Proteases are secreted by cells into the surrounding tissues to cause the destruction of proteins in extracellular material. • Secreted into an area (stomach) for the breakdown of protein in the diet. 6 Protease, peptidase, or proteinase? • Protease: is synonymous with peptidase. • Subclass EC 3.4.). 3 = hydrolase 4 = amide bond (peptide bond) • The Peptidases are comprised of two groups of enzymes: – Endopeptidases: cleave peptide bonds at points within the protein. – Exopeptidases: remove amino acids sequentially from either N or Cterminus. – Proteinase: synonymous with endopeptidase. 7 The EC nomenclature for proteases • • • • • • • • • • • • 3.4.11 Aminopeptidases3.4.13 Dipeptidases 3.4.14 Dipeptidyl-peptidases and tripeptidyl peptidases 3.4.15 Peptidyl-dipeptidases 3.4.16 Serine-type carboxypeptidases 3.4.17 Metallocarboxypeptidases 3.4.18 Cysteine-type carboxypeptidases 3.4.19 Omegapeptidases 3.4.21 Serine proteinases 3.4.22 Cysteine proteinases 3.4.23 Aspartic proteinases 3.4.24 Metallo proteinases 3.4.99 Proteinases of unknown mechanism 8 The four mechanistic classes of proteases • Serine proteases • Cysteine proteases • Aspartic proteases • Metallo proteases 9 Serine protease catalytic mechanism Serine Proteases often use the chymotrypsinogen numbering: His 57, Asp 102 and Ser 195 Two distinct evolutionarily unrelated families: (The classic example of convergent evolution) • Mammalian: chymotrypsin, trypsin or elastase • Bacterial :subtilisin. Ser/His/Asp Catalytic triad mechanism Ser: nucleophile His: general base Asp: help orient and neutralizes charge on His Covalent tetrahedral intermediates Covalent catalysis 10 Famous serine proteases • • • • • • • • • Trypsin This serine endopeptidase is the active form (MW 24,000) of the pancreatic proenzyme trypsinogen that is activated in the intestine by enterokinase. Chymotrypsin A group of serine proteases found in pancreatic secretions that hydrolyze peptide bonds in the process of digestion. This endopeptidase is made in the exocrine pancreas as a precursor that is activated by proteolytic cleavage in the duodenum by trypsin. Elastase MW 25.000. Serine protease of broad specificity made in the exocrine pancreas. Subtilisin Serine protease of bacterial origin. Plasmin Serine protease derived from the precursor plasminogen that breaks down insoluble fibrin thus disolving blood clots. Thrombin Serine protease present in plasma as a precursor called prothrombin (MW 72,000). The active form (MW 34,000) converts plasma fibrinogen into insoluble fibrin forming the basis of a blood clot. Kallikrein Causes the liberation of kinins from plasma protein precursors causing vasodilation and possibly hypotension. Streptokinase Protease produced by hemolytic bacteria that activates plasminogen thus producing plasmin which disolves fibrin. Therapeutically streptokinase has been used to dissolve pulmonary emboli and venous thromboses. Urokinase This protease is found in human blood and urine. It activates plasminogen by the cleavage of a propeptide forming the active serine protease plasmin. Therapeutically it is used to dissolve blood clots. 11 The Serine Proteases • • • • • Trypsin, chymotrypsin, elastase, thrombin, subtilisin, plasmin, TPA All involve a serine in catalysis - thus the name Ser is part of a "catalytic triad" of Ser, His, Asp Serine proteases are homologous, but locations of the three crucial residues differ somewhat Enzymologists agree, however, to number them always as His-57, Asp-102, Ser-195 Burst kinetics yield a hint of how they work! 12 13 Serine Protease Mechanism • • • • • A mixture of covalent and general acid-base catalysis Asp-102 functions only to orient His-57 His-57 acts as a general acid and base Ser-195 forms a covalent bond with peptide to be cleaved Covalent bond formation turns a trigonal C into a tetrahedral C The tetrahedral oxyanion intermediate is stabilized by N-Hs of Gly-193 and Ser-195 14 15 Artificial Substrate for Serine Proteases 16 17 Assay for Serine Proteases 18 Identification of Catalytic Residue Ser-195 19 20 21 Identification of Catalytic Residue His-57 (N-tosyl-L-phenylalanine chloromethylketone) 22 1. Conformational distortion forms the tetrahedral intermediate and causes the carboxyl to move close to the oxyanion hole 2. Now it forms two hydrogen bonds with the enzyme that cannot form when the carbonyl is in its normal conformation. 3. Distortion caused by the enzyme binding allows the hydrogen bonds to be maximal. 23 24 25 Serine proteases • Several different families - all have Ser in active site and all have the same reaction mechanism. • The two most commonly-studied are: • Trypsin family • Subtilisin family 26 Trypsin family of Serine Proteases • Includes: • trypsin • chymotrypsin (used for numbering) • elastase • thrombin • coagulation enzymes • plasmin • complement C1r and C1s 27 Serine proteases • Synthesized in zymogen (inactive) form and activated by cleavage of specific peptide bonds. • Catalytic triad: • • Serine 195 Histidine 57 Aspartate 102 • Numbering based on chymotrypsin. 28 Zymogen activation (Lehninger 8-31) 29 Chymotrypsin structure (Lehninger Fig 8-18) 30 Chymotrypsin Diagram from Branden & Tooze, 1991 31 Key residues • Ser 195 (1/28) labelled with DFP (diisopropylfluorophosphate) • His 57 labelled with Tosyl-Phechloromethyl ketone • Labelling of these residues inactivates serine proteases. 32 Protease - key areas • Binding • Main chain binding (non-specific) • Specificity pocket (residue 189) • Catalysis • active site - charge relay system • oxyanion hole formed 33 Specificity pocket explains protease specificity (residues 189, 216 and 226) Chymotrypsin aromatic Trypsin basic Diagram from Branden & Tooze, 1991 Elastase small, uncharged 34 Substrate modification (Lehninger) Small substrates for chymotrypsin 35 Two domains and other key residues Active site, S195, H57 and D102, lies between domains in chymotrypsin oxyanion hole (193195) main chain substrate binding (214-216) substrate specificity pocket (189,216,226) 36 Branden & Tooze, 1991 Oxyanion Hole Role • • • • Stabilizes the tetrahedral transition state Form H-bonds Uses catalytic triad plus Gly 193 Positions and orient substrate for hydrolysis • Stabilizes the TS 37 Zymogen to active A: zymogen Chymotrypsinogen B: active chymotrypsin 38 Subtilisin family (SB clan) • Includes furin, PC2 and PC3 - all active in prohormone processing • No sequence identity with trypsin family • Structure quite different (a/b structure; trypsin is b-barrel) • But - same reaction mechanism and organisation of catalytic triad 39 Serine protease families convergent evolution Trypsin family (H57, D102, S195) His Asp Ser Subtilisin family (D32, H64, S221) Asp His Ser 40 Chymotrypsin and subtilisin 41 Different structures but similar active sites Mechanism of Serine Protease His -57 H 102 Asp N O O N O 195 Ser N N H O O H C N C N O O H R2 H O H R2 R1 S1 R1 H O O N N 102 H O H H N C O R1 Tetrahedral Intermediate -1 O H H2O Oxyanion Hole P1 O H C O R1 Acyl-Enzyme H H N H O O H O H R2 N N O H N O O R2 N O C O O N N H O O R1 Tetrahedral Intermediate-2 C OH R1 P2 42 Cysteine protease catalytic mechanism Papain is the archetype and the best studied member of the family 1 4 Cysteine proteases sometimes use the Papain numbering system: Cys25 and His159. Asn175 helps stabilize the Cys- and His+ ion pair Unlike Ser protease Cys already ionized before substrate binding. a priori activated enzymes. 2 3 Cys25 and His159 play the same catalytic roles as Ser195 and His57 respectively in the serine proteases. Covalent tetrahedral intermediates 43 Famous cysteine proteases • Papain Thiol protease from the papaya. MW of about 23,400 used for tenderizing meats. • Bromelain Thiol protease found in pinapple juice and stem tissue. Glycoprotein of about 33,000 MW used in meat tenderizing, beer production and hydrolization of proteins in the food industry. • mammalian lysosomal Cathepsins (11) These intracellular cysteine proteases are involved in such diverse activities as blood clotting, cancer growth and metastasis and bone remodeling. Several varieties have been isolated from various tissues, each having specific functions. • cytosolic calpains (calcium-activated) • parasitic proteases (e.g Trypanosoma). • Interleukin-1-beta Converting Enzyme 44 Aspartic protease catalytic mechanism Bi-lobe enzymes with the active site located between two homologous lobes. 1 Asp general acid-base catalysis 2 "push-pull" mechanisms (steps 1 and 3) Non-covalently bound (neutral) tetrahedral intermediate. Low-barrier H-bonds may be involved. 3 45 Famous aspartic proteases • Pepsin Aspartic protease that cleaves bonds involving phenylalanine and leucine preferentially. MW 34,500. This endopeptidase is the principal digestive enzyme in gastric juice formed from the precursor pepsinogen (MW 38,000). (first protein crystals to diffract) • Renin Also known as angiotensinogenase. It is made by the juxtaglomerular cells in the kidney, released into the blood stream and acts to convert angiotensionogen into angiotensin I. It is an aspartyl protease having a molecular weight of about 40.000 Renin is elevated in some forms of hypertension. • cathepsins D • fungal proteases (penicillopepsin, rhizopuspepsin, endothiapepsin). • viral proteinases such as HIV protease (retropepsin). 46 The Aspartic Proteases • • • • Pepsin, chymosin, cathepsin D, renin and HIV-1 protease All involve two Asp residues at the active site Two Asps work together as general acid-base catalysts Most aspartic proteases have a tertiary structure consisting of two lobes (N-terminal and C-terminal) with approximate two-fold symmetry HIV-1 protease is a homodimer 47 48 Aspartic Protease Mechanism The pKa values of the Asp residues are crucial • One Asp has a relatively low pKa, other has a relatively high pKa • Deprotonated Asp acts as general base, accepting a proton from HOH, forming OHin the transition state • Other Asp (general acid) donates a proton, facilitating formation of tetrahedral intermediate 49 50 Asp Protease Mechanism • What evidence exists to support the hypothesis of different pKa values for the two Asp residues? • Bell-shaped curve is a summation of the curves for the two Asp titrations • In pepsin, one Asp has pKa of 1.4, the other 4.3 51 52 53 Mechanism of Aspartic protease O O 25`-Asp O H O O H C Proline N Aromatic Asp-25 O H H S1 O O 25`-Asp O H N Asp-25 HO O H O C R1 H R2 Tetrahedral Intermediate R2 H O O 25`-Asp OH N O H + OH C R1 Asp-25 O 54 HIV-1 Protease • • • • • A novel aspartic protease HIV-1 protease cleaves the polyprotein products of the HIV genome This is a remarkable imitation of mammalian aspartic proteases HIV-1 protease is a homodimer - more genetically economical for the virus Active site is two-fold symmetric Two Asp residues - one high pKa, one low pKa 55 56 Therapy for HIV? • • • • Protease inhibitors as AIDS drugs If the HIV-1 protease can be selectively inhibited, then new HIV particles cannot form Several novel protease inhibitors are currently marketed as AIDS drugs Many such inhibitors work in a culture dish However, a successful drug must be able to kill the virus in a human subject without blocking other essential proteases in the body 57 58 59 Metallo-protease catalytic mechanism • Most contain a Zn atom that is involved in catalysis. Sometimes Zn can be replaced by Co or Ni. • Zn is usually coordinated by 2 His and 1 Glu. Many contain the sequence HEXXH H • Non-covalent (oxyanion) tetrahedral intermediate. After the attack of a Zn-bound water molecule on the carbonyl group of the scissile bond. The intermediate breaks down by transfer of a proton From a Glu to the leaving group (not shown). 60 Famous metallo-proteases • Thermolysin Extracellular thermostable zinc metalloprotease from bacterial sources. • Collagenase Proteases that breakdown collagen that are produced by some species of bacteria such as Clostridium. Also isolated in human wound tissue, skin, bone, leukocytes, and cornea. Family of metalloproteases (MW 68,000125,000) that require zinc and calcium for activity. They are used in research for the isolation of cells from animal tissues. 61 Mechanism of Thermolysin His Glu 143 Glu His BH Zn2+ O C O H O H N C N C O H O Zn2+ His-231 R2 OH HO R1 OH HO O N H2 Zn2+ O H N R2 R1 H R1 Tetrahedral Intermediate C N C H R2 N Zn2+ C N N C N H O O H2O HO OH O C O H H N N H R2 62 Mechanism of Thermolysin • Zn2+ binds the amide carbonyl group • Zn2+ polarizes the carbon of carbonyl group • The nucleophile water (OH-) is activated by Glu-143 • His-231 is the acid for protonating the leaving group (R1-NH2) 63 The Schechter and Berger nomenclature for the description of protease subsites 64 Scissile bond and Subsite nomenclature solvent solvent P1’ P2 scissile bond P3 P1 I144 F84 D142 I86 V132 I101 I86 P87 S88 S90 K145 Y143 I144 L95 M91 S1 S3 S1 peptidase 65 Nucleophilic attack from Si vs. Re face P 1' N 7H P1' 8 O P 1' O 6 P1 si-face Ser/Lys protease P1 O N H N H P1 re-face Ser/His/Asp proteases 66 Angle of nucleophilic attack on a carbonyl The Dunitz-Bürgi angle (105°) From analysis of small molecule high resolution crystal structures it was observed that: The preferred O…C=O angle is 105° P1' O 105 From inhibitor and substrate co-crystal structures: The nucleophiles in the active-site of proteases (Ser hydroxyl Og, Cys Thiol Sg, waters) Are often orientated with respect to the scissile carbonyl close to the Dunitz-Bürgi angle (105°) N H O P1 Ser 67 Evolutionary Classification of proteases • MEROPS: Protease Data Base – Families: Proteases with statistically significant similarities in amino acid sequence. – Clans: Protease families that are thought to have common evolutionary origins. 68 Proteases as research tools • Limited proteolysis / sequencing / MS – Protein crystallization – Protein mobility (dynamics) – Protein-Protein interactions • NickPred: Web tool to understand limited proteolysis using PDBs • PROWL: A collection of protocols for limited proteolysis combined with Mass Spec 69 Mechanism of Electron Transfer in Biological System 1. Two One Electron Steps Mechanism 2. One Two Electron Steps Mechanism 70 Two One Electron Steps Mechanism • A free-radical intermediate must form which highly reactive and very energetic species • Discrete long-lived radical intermediates are infrequently seen in biological systems • Examples: flavin-Coenzyme • Vit C, E, K, CoQ • Metalloenzymes 71 One Two Electron Steps Mechanism 1. Hydride (H-:) Mechanism 2. H+ Abstraction Mechanism Most of enzyme hydrogenases are enzymatic dehydrogenation where H- ion transfer 72 One Two Electron Steps Mechanism Hydride Mechanism X Y H C H + X C HY + X C Carbonium Ion Proton Abstraction Mechanism X C H Y H + X C HY + X C Carbonanion Ion 73