* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Bacterial cultivation - Furry Helpers Pet Sitting
Survey
Document related concepts
Protein adsorption wikipedia , lookup
Citric acid cycle wikipedia , lookup
E. coli long-term evolution experiment wikipedia , lookup
Expanded genetic code wikipedia , lookup
Butyric acid wikipedia , lookup
Gaseous signaling molecules wikipedia , lookup
Phosphorylation wikipedia , lookup
Biosynthesis wikipedia , lookup
Metalloprotein wikipedia , lookup
Proteolysis wikipedia , lookup
Transcript
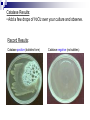
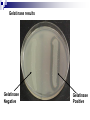
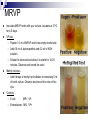
Lab Exercise: 15 Enzymes: 1. Catalase 2. Proteinase 3. MR-VP Enzymes Enzymes are important biosynthetic chemicals found in all organisms. The kinds of enzymes produced by an organism dictate the extent of its biosynthetic abilities. Enzymes are proteins that act as chemical catalysts (speeding up chemical reactions). The ability to produce enzymes is based in the genome (DNA) of the cell. If the cell has the appropriate genetic information for the synthesis of the enzyme then that enzyme is produced and the cell is able to use the specific reaction. Properties of enzymes include specificity and reusability. Enzymes Microorganisms, specifically bacteria, can be characterized by their ability to produce various enzymes. The presence of various enzymes in turn can be determined by the production of the appropriate end products in the medium (or their effects on the medium). Different bacteria produce various complement of enzymes and hence these can be useful in determining the identity of the bacterium based on the enzyme profile these organisms present. Catalase Bacteria vary in their ability to grow in the presence of atmospheric oxygen. The reason being that atmospheric oxygen (in the form it is usually found) is toxic to most cells. Only cells that have the ability to neutralize the toxic forms of oxygen are able to thrive in the presence of oxygen. Aerobes are those organisms that need and thrive in the presence of oxygen. (Obligate aerobes do not grow in the absence of oxygen) Anaerobes are those that cannot grow in the presence of oxygen (some are killed by oxygen) Facultative anaerobes are those organisms that are able to survive in the absence of oxygen but grow faster in the presence of oxygen. Catalase Catalase is the enzyme that breaks down toxic forms of oxygen (e.g. peroxides), neutralizing the toxic effect of those compounds. The presence of catalase indicates the microbe is capable of growing in the presence of oxygen (aerobe or facultative anaerobe), whereas the absence of catalase suggests the organism is anaerobic. The specific reaction catalyzed by catalase is: H2O2 ----------- H2O + O2 (gas) Under lab conditions the presence of catalase can be tested by adding liquid hydrogen peroxide drops directly onto the culture (cells/colony). If catalase is present in the organism then H2O2 is hydrolyzed and O2 is released as gas bubbles. Catalase Procedures • Controls: 1) Divide a DSA plate in half and 2) make a single streak with one control culture on each side of the plate. • Unknown: 1) Make a single streak on a DSA plate. • Incubate at 37° C overnight. Unknown DSA Controls (S.faecealis/ S.aureus) DSA Catalase Results: • Add a few drops of H2O2 over your culture and observe. Record Results: Catalase positive (bubbles form) Catalase negative (no bubbles) Amylase (Starch Hydrolysis) • Exoenzymes :enzymes that are produced by the cell and secreted outside to act on the substrate external to the cell. The broken down molecules can then be transported easily into the cell and used as nutrients by the cell. • Amylase :an exoenzyme that breaks down starch. • Reaction catalyzed by amylase: amylase Starch amylose + amylopectin Starch Amylase Procedures •Controls: 1) Divide a Starch Agar plate in half and 2) make a single streak with one control culture on each side of the plate. • Unknown: 1) Make a single streak on a Starch Agar plate. • Incubate at 37° C overnight. Unknown Controls (E.coli/ B. subtilus) Starch Agar Starch Agar Amylase Results •In the lab, the reaction can be visualized by addition of a Lugol’s iodine that will react with starch and produce blue/purple compound. • amylase + : a clearing in the blue zone around the growth indicates starch was broken down (amylase present). • amylase -: absence of a clearing in the blue indicates that starch is intact (no amylase). Controls (E. coli/ S.aureus) Unknown _ + Starch agar Starch agar Amylase: Results Negative for Amylase Positive for Amylase Gelatinase Gelatin is a complex polymeric protein molecule. Organisms that are able to produce the exoenzyme gelatinase are able to break down the gelatin structure and use it as a protein source (nutrient). Gelatin--polypeptides +free amino acids gelatinase Gelatinase The reaction can be made observable by the addition of chemicals that will react with the intact gelatin (unused) and form a white precipitate. The chemicals are combined in the form of a developer (mercuric chloride and hydrochloric acid), which precipitate the protein (gelatin) and form a white color. Absence of this white color around the area of growth indicate use of gelatin by the bacterium and hence a positive result for Gelatinase production by the organism. Absence of a clearing in the white precipitate formation indicates presence of intact gelatin (not used by bacterium) and hence a negative result for the gelatinase production by bacterium. Gelatinase Procedures •Controls: 1) Divide a Frazier’s Gelatin Agar plate in half and 2) make a single streak with one control culture on each side of the plate. • Unknown: 1) Make a single streak on a Frazier’s Gelatine Agar plate. • Incubate at 37° C overnight. Unknown Frazier’s Gelatine Agar Controls (E.coli/ B. subtilus) Frazier’s Gelatine Agar Gelatinase results Gelatinase Negative Gelatinase Positive Methyl Red Voges-Proskauer (MR-VP) test An important test to differentiate between glucose fermenting Enterobacteriaceae. Principle: Glucose is fermented to pyruvic acid by one of the two pathways: Fermentation of mixed acids Acetoin/ Naphtol+creatine pathway Methyl Red test Methyl Red is an acid-sensitive dye that is yellow at a pH above 4.5 and red at a pH below 4.5. When the dye is added to a culture of organisms growing in glucose broth, its color indicates whether the glucose has been broken down completely to highly acidic end products with a pH below 4.5 (methyl red positive = red), or only partially to less acidic end products with a pH above 4.5 (methyl red negative = yellow). Voges-Proskauer test The Vogues-Proskauer test can be performed on the same glucose broth culture used for the methyl red test (MRVP broth). One of the glucose fermentation end products produced by some organisms is a Naphtol+creatine. The VP reagents (alphanaphthol and potassium hydroxide solution) oxidize this compound having a pink to red color. VP-positive organisms are those reacting in the test to give this pink color change. MRVP Inoculate MRVP broth with your culture. Incubate at 37oC for 4-5 days. VP test- Pipette 1.0 ml of MRVP broth into empty sterile tube. Add 0.6 ml of alphanaphthol and 0.2 ml of KOH solution. Shake the tube well and allow it to stand for 10-20 minutes. Observe and record the color. Methyl red test - Add 5 drops of methyl red indicator to the remaining 5 ml of broth culture. Observe and record the color of the dye. (Methyl red test) Record results of previous lab Catalase Controls: S. faecalis, S. aureus unknown Gelatinase Controls: E.coli, B. subtilis, unknown MRVP E. coli, Enterobacter aerogenes Unknown MRVP Inoculate MRVP broth with your culture. Incubate at 37oC for 4-5 days. VP test- Pipette 1.0 ml of MRVP broth into empty sterile tube. Add 0.6 ml of alphanaphthol and 0.2 ml of KOH solution. Shake the tube well and allow it to stand for 10-20 minutes. Observe and record the color. Methyl red test – Add 5 drops of methyl red indicator to remaining 5 ml of broth culture. Observe and record the color of the dye. Controls E.coli: Enterobacter: MR-, VP+ MR+, VP- Enzymes II Lab 16 SIM test- Sulfide, Indole, Motility Indole is a by-product of the metabolic breakdown of the amino acid tryptophan used by some microbes. The presence of indole in a culture grown in a medium containing tryptophan can be readily demonstrated by adding Kovac's reagent to the culture. If indole is present, it combines with the reagent to produce a brilliant red color. If it is not present, there will be no color except that of the reagent itself. This test is of great value in the battery used it identify enteric bacteria. * RUN THE INDOLE PART OF THE TEST FIRST SIM Hydrogen sulfide is produced when amino acids containing sulfur are metabolized by microbes. Hydrogen sulfide formed during growth combines with the metallic ions (iron) to form a metal sulfide that blackens to medium. SIM- motility Motility - This is a tubed semisolid agar that can be used also to demonstrate motility. It is inoculated by stabbing the wire needle straight down the middle of the agar and withdrawing along the same path. Motile organisms will migrate through the agar, while non-motile microbes will only grow along the stab line SIM- Sulfide, Indole, Motility tests Procedure Citrate test The ability of some organisms, such as Enterobacter aerogenes and Salmonella typhimurium to utilize citrate as a sole source of carbon. Citrate is a simple carbohydrate and the test is useful differentiation characteristic in working with intestinal bacteria. Koser's citrate medium and Simmons citrate agar are two media used to detect this ability in bacteria. Both of these are synthetic media in which sodium citrate is the sole source of carbon, and nitrogen is supplied by ammonium salts instead of amino acids. Simmons citrate agar contains the indicator bromthymol blue which changes from green to blue when growth of organisms causes alkalinity. Citrate test Inoculate a slant of Simmons Citrate agar with your culture, by streaking the surface. Incubate at 37°C for 1-2 days. E.coli, and K. pneumonia Unknown Examine the slant. Negative- If the organism did not grow the slant will remain green. E. coli Positive- If the organism did grow, the slant will be partly or completely blue in color. K. pneumonia Urease reaction Urea is a protein compound. Bacteria that are able to break down the molecule have access to other protein nutrients. Bacteria that have the enzyme urease are able to use the urea. Urea------------- CO2 +NH3 The reaction is made observable by including a pH indicator in the medium. Phenol red is the indicator (is peach color at neutral pH, turns yellow below 6.8 and pink above 8.4). Urease test Exercise- inoculate E.coli, Proteus vulgais unknown Incubate 48 h at 37 0c Interpretation: The release of ammonia by the breakdown of urea results in the an alkaline pH of the medium which will turn the medium pink. Thus pink color indicates production of urease by the organisms Absence of pink color indicates a negative test for urease. Carbohydrate utilizationTSI, fermentation tubes (F-tubes) Lab 17 Carbohydrate utilization Carbohydrates are sugars and they serve as energy source for bacteria Bacteria use metabolic pathways to degrade sugars and generate energy (ATP) in aerobic conditions. In anaerobic conditions some bacteria can switch to fermentation of sugars with only small gain of ATP. The ability of cells to catabolize specific substrates is dependent on their ability to produce appropriate enzymes. Fermentation Fermentation is the catabolism of sugars in the absence of oxygen where the final electron acceptors is an organic molecule. Fermentation typically results in the formation of organic acids and alcohols that will accumulate in the medium. This will result in the lowering of the pH of the medium which can be visualized by the addition of an indicator in the media. Gas production-Some bacteria characteristically produce gases during the fermentation process, which can be made visible by the addition of inverted tubes (F-tubes) in the case of liquid media and cracks in the agar in solid medium. F-tubes (Fermentation tubes) The ability to ferment a specific sugars is dependant on the presence of the enzymes required for the transport and metabolism of that sugar. Thus fermentation of various sugars can be used to characterize bacteria. The F-tubes use phenol red in the medium as pH indicator and the use of inverted tubes to detect production of gases. Results are recorded as Acid / Acid Gas / Alkaline / No reaction. KIA – Kligler Iron Agar KIA is a medium used in the identification of Gram-negative enteric rods. KIA contains glucose and lactose, but there is 10 times more lactose than glucose. KIA contains a pH-sensitive color indicator. Fermentation of these sugars can be determined by color change of the phenol red indicator, from red to yellow for acid production. Principles of the Procedure KIA contains: Protein source (beef extract Two sugars (dextrose 0.1% and lactose 1% ) phenol red for detecting carbohydrate fermentation ferrous ammonium sulfate for detection of hydrogen sulfide production (indicated by blackening in the butt of the tube). The KIA has a two reaction chamber: Upper/slant part- aerobic –oxidative decarboxylation of amino acids Lower/ Butt anaerobic slant butt Reactions tested by KIA test Fermentation of sugars – acid production lower the pH (yellow) Proteins catabolism -broken down resulting in alkaline products, which increase the pH (pink) H2S production Sugar fermentation / protein metabolismas a source of energy- protein sparing. Cells will prefer to use sugars for energy when they are present, sparing the proteins; Proteins will be used only after the sugar source has been depleted. Protein metabolism results in alkaline end products and an increased pH of the media. When a bacterium ferments the sugar and lowers the pH and depletes all the sugars. The proteins in the medium will then be broken down resulting in alkaline products, which increase the pH and reverse the results of sugar fermentation. Thus fermentation reactions have to be read within 18-24 hours before sugar reversion can occur. H2S production Hydrogen sulfide (H2S) production, blackens the medium if it occurs. The H2S reacts with iron in the medium to produce FeS, which is black. KIA- procedure Using a straight inoculating wire, stab the butt and streak the surface of agar slant. Do not close lid tightly. Incubate 1-2 days 37oC. Observe color of butt and slant as described above. Was there any H2S production? Is the strain motile? This can be determined by the grow pattern in the stab. Record all results. Interpretation in following chart: KIA- Interpretation butt / slant Interpretation yellow / acid yellow glucose and lactose and/or sucrose are fermented yellow / acid orange-red glucose only fermented or pink = alkaline orange-red/ neutral yellow acid glucose only utilized, aerobically orange-red / neutral orange-red neutral no fermentation bubbles cracks - gas production black - hydrogen sulfide produced Interpretation 1 2 3 GAS 4 5 KIA test - Results 6 NOTES: • Red slant = peptone catabolization •Yellow butt = glucose fermentation only •Yellow slant = glucose and lactose ferm. •Black butt = glucose ferm and H2S prod. C - K/K - control, no inoculation, no change 1- K/K - no sugar fermentation, red slant due to peptone catabolization 2- K/A - glucose only fermentation, facultative anaerobe, no gas 3- K/A/H2S - glucose fermentation, fac. anaerobe, H2S gas production 4- A/AG - glucose and lactose fermentation, gas production 4A- A/A-KG - like #4, but peptone catabolism leadd to pink on slant 5- A/A/H2S - like #4, but the gas produced is H2S 6- A/K - aerobic, capable of glucose and lactose fermentation












































![fermentation[1].](http://s1.studyres.com/store/data/008290469_1-3a25eae6a4ca657233c4e21cf2e1a1bb-150x150.png)