* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Lab 7: Mutation, Selection and Drift
Saethre–Chotzen syndrome wikipedia , lookup
Quantitative trait locus wikipedia , lookup
SNP genotyping wikipedia , lookup
Human genetic variation wikipedia , lookup
BRCA mutation wikipedia , lookup
Skewed X-inactivation wikipedia , lookup
Group selection wikipedia , lookup
Polymorphism (biology) wikipedia , lookup
Human leukocyte antigen wikipedia , lookup
Koinophilia wikipedia , lookup
Hardy–Weinberg principle wikipedia , lookup
Frameshift mutation wikipedia , lookup
Point mutation wikipedia , lookup
Dominance (genetics) wikipedia , lookup
Microevolution wikipedia , lookup
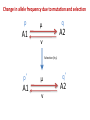
Lab 10: Mutation, Selection and Drift Goals 1. Effect of mutation on allele frequency. 2. Effect of mutation and selection on allele frequency. 3. Effect of mutation, selection and drift on allele frequency. 4. Compare mutation rates in different groups. Change in allele frequency due to mutation p μ A1 v q p q q A2 Change in allele frequency due to mutation q p q Where, µ = rate of forward mutation (From A1 to A2) ν = rate of backward mutation (From A2 to A1) At equilibrium, ∆q=0, μ qeq μ ν Assumptions: 1. Large population size. 2. No selection. Problem 1. Eye color in humans was once believed to be controlled by a single gene, with the brown eye allele being the dominant wild-type. Recent studies, however, revealed that eye color is actually a polygenic trait. Although 74% of the variation for eye color is determined by the Eye Color 3 (EYCL3) locus located on chromosome 15 (with most variation explained by only 3 single nucleotide polymorphisms!), there are at least two other loci with weaker but significant control over this trait. The frequency of the recessive allele associated with blue eyes (let this be allele A2) is 0.83 in Europeans. Assuming that an equilibrium has been reached between forward and backward mutation, and the rate of forward mutation (i.e., from wild-type A1 alleles to A2 alleles) is μ = 10-6, calculate the rate of backward mutation (i.e., from A2 to A1). (5 minutes) Given, A1- Brown eye ; A2- Blue Eye; A1>A2, qeq = 0.83 μ (A1 to A2) = 10-5 ν(A2 to A1) = ? Change in allele frequency due to mutation and selection p A1 q μ A2 v Selection (h,s) p’ A1 μ v q’ A2 Change in allele frequency due to mutation and selection Case 1: When ν ≈ 0; population size is large and h=0 (A1 completely dominant to A2): q eq s Case 2: When ν ≈ 0; population size is large and h>>0 : q eq hs Problem 2. Sequence analysis of EYCL3 reveals that this locus is likely to have been under very strong selection, and the allele associated with blue eyes (as well as with light brown hair and pale skin color) is likely to have been favored in Europe, but not in Africa and East Asia. Let us assume that melanoma (skin cancer, which is more likely to develop in people with light skin color) reverses the direction of selection and the blue eye/light skin allele now becomes selected against with s = 0.12. Calculate the equilibrium value of q (the frequency of the blue eye allele A2) in an infinitely large population if the rate of forward mutations is μ = 10-6, the rate of backward mutations is ν = 0, and if: a. A1 is completely dominant to A2. b. There is additivity. c. If the equilibrium frequencies of A2 in a) and b) are different, explain why. If not, explain why not. d. Under which type of dominance is equilibrium approached faster and why? Provide output from Populus to support your calculations. In Populus, use value of s, h, and μ and for enough # of generations with six initial allele frequency. Mutation, selection and genetic drift p A1 q μ A2 v Drift xij P(Yt 1 i | Yt j ) (2 N )! p'i q'2 N i (2 N i)!i! p’ A1 Selection (h,s) μ v q’ A2 Mutation, selection and genetic drift Mean frequency of A2 (harmful mutant) allele, when h=0 and Ne and ν are very small : 2N e q s Mean frequency ofA2 allele, when h >> √µs and ν is very small: q hs Problem 3. Starting with the conditions in Problem 2, calculate the expected value of q in a finite population if: a. A1 is completely dominant to A2 and Ne = 50. b. Same as a) but with Ne = 10,000. c. There is additivity, and Ne = 50. d. Same as c) but with Ne = 10,000. How does Ne affect the mean frequency of A2 with each type of dominance? Under which level of dominance does Ne have a stronger effect? Why? Provide output from PopG to support your interpretation.(15 minutes). Comparing mutation rates in different groups Mutation rates can be estimated directly by comparing the genotypes of parents to those of their offsprings. Sometimes it is of great interest to compare rates of mutation between different groups (e.g., between a group exposed to radiation and a control group). This can be done using contingency table tests. Two methods to analyze contingency tables are the Chi-square test and Fisher’s exact test. Example: Group 1(Control) Group 2 (Exposed group) No. of No. of alleles No. of mutations tested mutations No. of alleles tested 3 60 50 5 (Oi Ei ) Ei i 1 4 2 2 df = (number of rows − 1)(number of columns − 1) = 1 2x2 contingency table and χ2 test Om Ou gr1 3 47 gr2 5 55 Em Eu (50*8)/110= (50*102)/110 3.63 = 46.36 (60*8)/110= (60*102)/110 4.36 = 55.63 ((Om-Em)^2)/Em ((Ou-Eu)^2)/Eu 0.111 0.008 0.092 0.007 0.204 0.016 chi square 0.2202 Here, the calculated χ2 value (0.2202) is less than the critical value (3.8415) for α=0.05. Hence we fail to reject null hypothesis i.e. there is no significant difference in mutation rates of the two groups. Problem 4. The following numbers of mutations have been observed in minisatellite loci of humans exposed to radiation in Chernobyl compared to a control group: Use the Chi-square test to determine whether mutation rates differ significantly between: a. Alleles inherited from the mother and alleles inherited from the father in the control group. b. Alleles inherited from the mother and alleles inherited from the father in the exposed group. c. The control and the exposed groups for alleles inherited from the mother. d. The control and the exposed groups for alleles inherited from the father. e. GRADUATE STUDENTS ONLY: Repeat all tests using Fisher’s exact test. Control No. of No. of alleles mutations tested Exposed No. of No. of alleles mutations tested Alleles inherited from mother 9 721 26 1650 Alleles inherited from father 26 827 141 1701