* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Slides
Signal transduction wikipedia , lookup
Cell culture wikipedia , lookup
Extracellular matrix wikipedia , lookup
Cell encapsulation wikipedia , lookup
Cellular differentiation wikipedia , lookup
Organ-on-a-chip wikipedia , lookup
List of types of proteins wikipedia , lookup
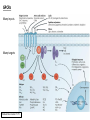
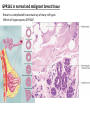
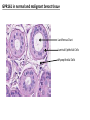
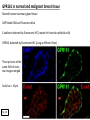
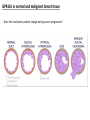
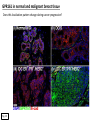
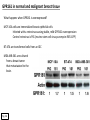
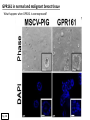
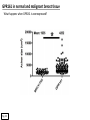
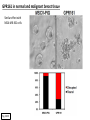
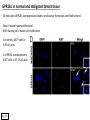
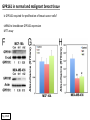
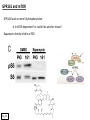
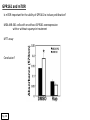
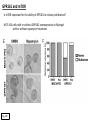
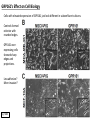
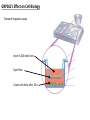
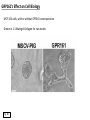
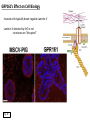
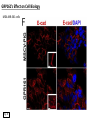
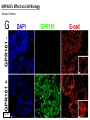
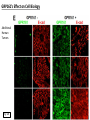
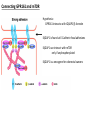
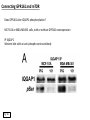
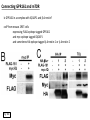
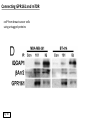
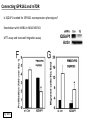
G-protein–coupled receptor GPR161 is overexpressed in breast cancer and is a promoter of cell proliferation and invasion Michael E. Feigin, Bin Xue, Molly C. Hammell, and Senthil K. Muthuswamy Cold Spring Harbor Laboratory Stony Brook University, NY University of Toronto Proceedings of the National Academies of Science, USA March 18, 2014 111:491-496 Triple-Negative Breast Cancer (TNBC) No expression of • Estrogen Receptor (ER) • Progesterone Receptor (PR) • ErbB2 (EGF Receptor/HER2) ~25% of all breast cancers Generally worse prognosis and lack of targeted therapies Tamoxifen family of drugs targets ER Herceptin targets HER2 To develop new therapies for TNBC, we need to understand its causes. Triple-Negative Breast Cancer (TNBC) ~15% of TNBCs are associated with BRCA1 or BRCA2 mutations These gene account for about most of familial breast cancers, ~5-10% of total Both are involved with DNA repair Origin of BRCA+ TNBCs is unclear Analyzed patient tumor genomes in The Cancer Genome Atlas (TCGA) Specifically looked for overexpressed G Protein Coupled Receptors (GPCRs) TGCA Screenshot GPCRs Seven transmembrane domains Receptors for many extracellular signals Leads to the activation of a heterotrimeric G protein Humans encode ~800 GPCRs (~4% of all genes!) Very “druggable” ~30% of current drugs target a GPCR agonists or antagonists GPCRs Many inputs Many targets Nature Rev. Cancer 7:79 GPR161 in TNBC Seeking GPCRs that are overexpressed in TNBC Can’t just look at genomic DNA sequence! Seeking a change in expression not a mutation Looked at RNA sequencing data (RNA-seq) at TGCA cDNAs generated from tumor mRNA and sequenced 98 TNBC compared to 100 normal breast tissue samples (nonmatched) Followed 366 GPCRs (all known to not be involved with the senses) Seeking GPCRs that are over-represented (on enriched) in the RNA-seq data 45 GPCRs were upregulated significantly (at least 2-fold) GPR161 in TNBC GPR161 is upregulated 2.2-fold in TNBC Not upregulated in ER+ tumors (LumA/B) HER2+ tumors Fig. 1A GPR161 in TNBC GPR161 is upregulated 2.2-fold in TNBC GPR161 is upregulated in other breast cancer datasets: Richardson Breast 2 Panel (40 samples) Farmer Breast Study (49 samples) Fig. 1A, S1AB GPR161 in TNBC For most of these samples, clinical data are available on the patient. Did high levels of GPR161 expression correlate with relapse-free survival rates? Compared highest and lowest quartile of GPR161 expression Among basal type TNBC, high GPR161 expression decreased time to relapse by 113% for lymph node positive and 54% for all basal cancers Fig. 1BC GPR161 in TNBC For any type of TNBC, high GPR161 expression decreased time to relapse by 27% Fig. S1C GPR161 in normal and malignant breast tissue Breast is a complicated tissue made up of many cell types. Which cell types express GPR161? GPR161 in normal and malignant breast tissue Lactiferous Duct Luminal Epithelial Cells Myoepithelial Cells GPR161 in normal and malignant breast tissue Normal human mammary gland tissue DAPI binds DNA and fluoresces blue E-cadherin detected by fluorescent IHC (marker for luminal epithelial cells) GPR161 detected by fluorescent IHC (using a different fluor) Three pictures of the same field of view; two images merged Scale bar = 10mm Fig. 1D GPR161 in normal and malignant breast tissue Does this localization pattern change during cancer progression? GPR161 in normal and malignant breast tissue Does this localization pattern change during cancer progression? Fig. 1E GPR161 in normal and malignant breast tissue What happens when GPR161 is overexpressed? MCF-10A cells are immortalized breast epithelial cells Infected with a retrovirus causing stable, mild GPR161 overexpression Control retrovirus is PIG (murine stem cell virus puromycin-IRES-GFP) BT-474 are transformed cells from an IDC MDA-MB-361 are cultured from a breast tumor that metastasized to the brain. Fig. 2A GPR161 in normal and malignant breast tissue What happens when GPR161 is overexpressed? MCF-10A cells can be grown in 3D, leading to ducts. Plastic plates are coated with Matrigel – extracellular matrix proteins secreted by a cell line Lots of collagen, laminin, entactin and some growth factors Cultured for two weeks Fig. 2B GPR161 in normal and malignant breast tissue What happens when GPR161 is overexpressed? Fig. 2B GPR161 in normal and malignant breast tissue What happens when GPR161 is overexpressed? Fig. 2C GPR161 in normal and malignant breast tissue Similar effect with MDA-MB-361 cells Fig. S1DE GPR161 in normal and malignant breast tissue So how does GPR161 overexpression lead to multiacinar formation and filled lumens? Does it cause hyperproliferation? Cells cultured in 96-well plate followed by MTT assay GPR161 in normal and malignant breast tissue So how does GPR161 overexpression lead to multiacinar formation and filled lumens? Does it cause hyperproliferation? Cells cultured in 96-well plate followed by MTT assay Each cell line was normalized to its control Fig. 2E GPR161 in normal and malignant breast tissue So how does GPR161 overexpression lead to multiacinar formation and filled lumens? Does it cause hyperproliferation? Ki67 staining as a marker of proliferation In controls, Ki67+ cells in 6.3% of acini. In GPR161 overexpressors, Ki67+ cells in 57.5% of acini. Fig. 2D GPR161 in normal and malignant breast tissue Is GPR161 required for proliferation of breast cancer cells? shRNA to knockdown GPR161 expression MTT assay Fig. 2FGH GPR161 and mTOR What pathway(s) is used by GPR161 to affect proliferation? Reverse Phase Protein Array (RPPA) data from TGCA Proteins from various cancers plated as an array Incubated with a specific antibody Quantify differences across tumors GPR161 and mTOR What pathway(s) is used by GPR161 to affect proliferation? Reverse Phase Protein Array (RPPA) data from TGCA Many alterations, including phospho-EIF4BP1 and phospho-RPS6KA1 and others Fig. 3A GPR161 and mTOR Many of these proteins are in the mTOR pathway Connects nutrient level and growth control GPR161 and mTOR Examined phosphorylation state of key proteins in MDA-MB-361 cells with or without GPR161 overexpression Fig. 3B GPR161 and mTOR GPR161 leads to more S6 phosphorylation. Is it mTOR-dependent? or could it be another kinase? Rapamycin directly inhibits mTOR Fig. 3C GPR161 and mTOR Is mTOR important for the ability of GPR161 to induce proliferation? MDA-MB-361 cells with or without GPR161 overexpression with or without rapamycin treatment MTT assay Conclusion? Fig. 3D GPR161 and mTOR Is mTOR important for the ability of GPR161 to induce proliferation? MCF-10A cells with or without GPR161 overexpression in Matrigel with or without rapamycin treatment Fig. 3EF GPR161 and mTOR Is GPR161 upstream or downstream of mTOR? Fig. 3 GRP161’s Effect on Cell Biology Cells with elevated expression of GPR161 just look different in subconfluent cultures. Controls formed colonies with rounded edges. GPR161 overexpressing cells showed sharp edges and projections. Less adhesive? More invasive? Fig. S2BC GRP161’s Effect on Cell Biology Transwell migration assay Insert 5,000 cells here 8mm filter Count cells here after 24 h GRP161’s Effect on Cell Biology Transwell migration assay Two cell lines, with or without GPR161 overexpression Fig. 4A GRP161’s Effect on Cell Biology MCF-10A cells, with or without GPR161 overexpression Grown in 1:1 Matrigel:Collagen for two weeks Fig. 4B GRP161’s Effect on Cell Biology Invasive cells typically down-regulate Laminin-V Laminin-V detected by IHC in red structures are “disrupted” Plasma Membrane Fig. 4C GRP161’s Effect on Cell Biology Cell-cell adhesion is mediated by E-Cadherin (among many other proteins) Measure E-Cadherin levels in MCF-10A cells “modestly reduced” Fig. 4E GRP161’s Effect on Cell Biology Concanavalin A (Con A) is a plant protein that binds certain carbohydrate groups that are abundant on glycoproteins and glycolipids Total Cell Lysase (TCL) was run over ConA-beads removes most plasma membrane fragments intracellular membranes remain Suggests E-cadherin is significantly mislocalized Fig. 4E GRP161’s Effect on Cell Biology MDA-MB-361 cells Fig. 4F GRP161’s Effect on Cell Biology Human Tumors Fig. 4G GRP161’s Effect on Cell Biology Additional Human Tumors Fig. S2E Connecting GPR161 and mTOR GPR161 We concluded that mTOR is downstream of GPR161. But how are they connected? ? Connecting GPR161 and mTOR Hypothesis: GPR161 interacts with IQGAP1/b-Arrestin IQGAP1 is found at E-Cadherin focal adhesions IQGAP1 can interact with mTOR only if unphosphorylated IQGAP1 is a oncogene for colorectal cancers Connecting GPR161 and mTOR Does GPR161 alter IQGAP1 phosphorylation? MCF-10A or MDA-MB-361 cells, with or without GPR161 overexpression IP IQGAP1 Western blot with an anti-phospho-serine antibody Fig. 5A Connecting GPR161 and mTOR Is GPR161 in a complex with IQGAP1 and b-Arrestin? coIP from mouse 293T cells expressing FLAG-epitope tagged GPR161 and myc-epitope tagged IQGAP1 and sometimes HA-epitope tagged b-Arrestin-1 or b-Arrestin-2 Fig. 5BC Connecting GPR161 and mTOR coIP from breast cancer cells using untagged proteins Fig. 5D Connecting GPR161 and mTOR Is IQGAP1 needed for GPR161 overexpression phenotypes? Knockdown with shRNA in MDA-MB-361 MTT assay and transwell migration assay Fig. 5EFG Connecting GPR161 and mTOR Both GPR161 and IQGAP1 have been reported to be overexpressed in breast tumors. Of 748 breast tumors in TGCA 166 (22.2%) had amplified GPR161 39 (5.2%) had amplified IQGAP2 Are the same tumors overexpressing both genes? Or do breast tumors overexpress one gene and not the other? 13 show overexpression of both Much more than expected by chance Fig. 5H