* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download neutphylo
Maximum parsimony (phylogenetics) wikipedia , lookup
Dual inheritance theory wikipedia , lookup
DNA barcoding wikipedia , lookup
Transitional fossil wikipedia , lookup
Adaptive evolution in the human genome wikipedia , lookup
Point mutation wikipedia , lookup
Viral phylodynamics wikipedia , lookup
Computational phylogenetics wikipedia , lookup
Population genetics wikipedia , lookup
Quantitative trait locus wikipedia , lookup
Selection in Nature
I.
Ecological Interactions
II. Constraints on Selection
III. Back to the Neutral Theory!
III. Back to The Neutral Theory
A. Neutral Variation
- change in protein that does not affect fitness
- ‘silent’ or ‘synonymous’ mutations are the prototype
III. Back to The Neutral Theory
A. Neutral Variation
- change in protein that does not affect fitness
- ‘silent’ or ‘synonymous’ mutations are the prototype
B. Predictions and Results
1. Rates of molecular evolution should vary in functional and nonfunctional regions
III. Back to The Neutral Theory
A. Neutral Variation
- change in protein that does not affect fitness
- ‘silent’ or ‘synonymous’ mutations are the prototype
B. Predictions and Results
1. Rates of molecular evolution should vary in functional and nonfunctional regions
- Rates should vary in different codon positions. Variation at the
third position should be higher, because these are usually silent mutations.
Mutations at the second position change amino acids, and these changes are
deleterious. PATTERN CONFIRMED.
- Rates should vary in coding and non-coding regions. Variation in
Introns should occur more rapidly than variation in exons, since introns are
not transcribed and are also invisible to selection. PATTERN CONFIRMED
- Rates should vary in functional and non-functional regions of
proteins. PATTERN CONFIRMED
III. Back to The Neutral Theory
A. Neutral Variation
- change in protein that does not affect fitness
- ‘silent’ or ‘synonymous’ mutations are the prototype
B. Predictions and Results
1. Rates of molecular evolution should vary in functional and nonfunctional regions
2. Rates of replacement (substitution of one fixed allele by another
that reaches fixation) should be constant over geologic time.
III. Back to The Neutral Theory
A. Neutral Variation
- change in protein that does not affect fitness
- ‘silent’ or ‘synonymous’ mutations are the prototype
B. Predictions and Results
1. Rates of molecular evolution should vary in functional and nonfunctional regions
2. Rates of replacement (substitution of one fixed allele by another
that reaches fixation) should be constant over geologic time.
- If changes are random and mutations occur at a given rate, then
replacement should "tick" along like a clock.
- Selection should speed rates when a new adaptive combination
occurs, like in obviously adaptive morphological trait. Then inhibit further
change unless it is adaptive or neutral - PATTERNS CONFIRMED (usually).
III. Back to The Neutral Theory
A. Neutral Variation
- change in protein that does not affect fitness
- ‘silent’ or ‘synonymous’ mutations are the prototype
B. Predictions and Results
1. Rates of molecular evolution should vary in functional and nonfunctional regions
2. Rates of replacement (substitution of one fixed allele by another
that reaches fixation) should be constant over geologic time.
3. Rates of morphological change should be independent of the rate
of molecular change.
III. Back to The Neutral Theory
A. Neutral Variation
- change in protein that does not affect fitness
- ‘silent’ or ‘synonymous’ mutations are the prototype
B. Predictions and Results
1. Rates of molecular evolution should vary in functional and nonfunctional regions
2. Rates of replacement (substitution of one fixed allele by another
that reaches fixation) should be constant over geologic time.
3. Rates of morphological change should be independent of the rate
of molecular change.
- "Living Fossils" show extreme genetic change and variation, yet
have remained morphologically unchanged for millennia. And, their rate of
genetic change in this morphologically constant species is the same as in
hominids, which have changed dramatically in morphology over a short
period. PATTERN CONFIRMED
III. Back to The Neutral Theory
A. Neutral Variation
- change in protein that does not affect fitness
- ‘silent’ or ‘synonymous’ mutations are the prototype
B. Predictions and Results
1. Rates of molecular evolution should vary in functional and nonfunctional regions
2. Rates of replacement (substitution of one fixed allele by another
that reaches fixation) should be constant over geologic time.
3. Rates of morphological change should be independent of the rate
of molecular change.
4. A truly neutral clock should tick off mutations at a constant rate.
But should this ticking occur per unit time, or per generation? (Selection will
proceed faster in organisms with short generation times…)
III. Back to The Neutral Theory
A. Neutral Variation
- change in protein that does not affect fitness
- ‘silent’ or ‘synonymous’ mutations are the prototype
B. Predictions and Results
1. Rates of molecular evolution should vary in functional and nonfunctional regions
2. Rates of replacement (substitution of one fixed allele by another
that reaches fixation) should be constant over geologic time.
3. Rates of morphological change should be independent of the rate
of molecular change.
4. A truly neutral clock should tick off mutations at a constant rate.
But should this ticking occur per unit time, or per generation?
- Mutations occur during DNA replication of the DNA, so a truly
neutral clock should tick at a rate dependent on the generation time of the
organism. Species with rapid generation times should accumulate mutations
at a faster rate than long-lived species with slower generation times.
- This is true of non-coding DNA... but not true for proteins. Proteins
accumulate mutations in absolute time, not generational time. THIS IS
INCONSISTENT WITH THE NEUTRAL MODEL
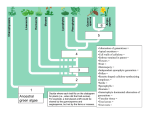
Rate of evolution in proteins
Clustering analysis based on amino acid
similarity across seven proteins from 17
mammalian species.
Rate of evolution in proteins
Now, we date the oldest mammalian fossil,
which our evolution hypothesis dictates
should be ancestral to all mammals, both the
placentals (species 1-16) and the marsupial
kangaroo. …. This dates to 120 million years
16
Rate of evolution in proteins
And, through our protein analysis, we already
know how many genetic differences
(nitrogenous base substitutions) would be
required to account for the differences we see
in these proteins - 98.
16
Rate of evolution in proteins
So now we can plot genetic change against time.
16
Rate of evolution in proteins
Repeat for every node. How old is the putative common ancestor
between rabbits and rodents (58 mya). How many substitutions
required to explain the differences in AA sequence? (50).
Rate of evolution in proteins
Just where genetic analysis of two different EXISTING species
predicts.
16
Rate of evolution in proteins
And all 16 nodes? Describes a straight line (constant
mutation rate).
III. Back to The Neutral Theory
A. Neutral Variation
B. Predictions and Results
C. Ohta’s “Nearly Neutral” Model
III. Back to The Neutral Theory
A. Neutral Variation
B. Predictions and Results
C. Ohta’s “Nearly Neutral” Model
- Included weak selection against slightly
deleterious alleles. if s < 1/2Ne, then alleles are
essentially neutral and become fixed as drift
would predict.
- In small populations, drift predominates
unless selection is fairly strong (in a
population of Ne = 5, drift will predominate
unless s > 0.1).
- In large populations, selection predominates,
even if it is fairly weak (if Ne = 10,000, then
selection will predominate if s > 0.00005).
SO.
Sub. Rate
- We observe a constant AA substitution rate across species, even
though we would expect that species with shorter generation times should
have FASTER rates of substitution.
OBS.
EXP.
Short
GEN TIME
Long
SO.
- We observe a constant AA substitution rate across species, even
though we would expect that species with shorter generation times should
have FASTER rates of substitution.
- So, something must be 'slowing down' this rate of substitution in
species with short gen. times. What's slowing it down is their large
populations size, such that the effects of drift, alone, are reduced.
Sub. Rate
LARGE
POP. SIZE
OBS.
EXP.
Short
GEN TIME
Long
SO.
- We observe a constant AA substitution rate across species, even
though we would expect that species with shorter generation times should
have FASTER rates of substitution.
- So, something must be 'slowing down' this rate of substitution in
species with short gen. times. What's slowing it down is their large
populations size, such that the effects of drift, alone, are reduced.
- Likewise, species with long generation times have small
populations, and substitution by drift and fixation is more rapid than
expected based on generation time, alone.
Sub. Rate
SMALL POP.
SIZE
OBS.
EXP.
Short
GEN TIME
Long
SO.
- The constant rate of AA substitution across species is due to the
balance between the effects of generation time and population size.
Sub. Rate
SMALL POP.
SIZE
OBS.
EXP.
Short
GEN TIME
Long
III. Back to The Neutral Theory
A. Neutral Variation
B. Predictions and Results
C. Ohta’s “Nearly Neutral” Model
D. New Developments
…. Some ‘synonymous’ substitutions are NOT neutral!
- ‘synonymous’ mutations in exons may slow the rate of protein
synthesis and cell growth.
Patrick Goymer (2007) 'Genetic variation: Synonymous mutations break
their silence', Nature Reviews Genetics 8, 92 (February 2007).
Chamary, J. V. & Parmley, J. L. & Hurst, L. D. 'Hearing silence: non-neutral
evolution at synonymous sites in mammals'. Nature Rev. Genet. 7, 98-108
(2006).
Grzegorz Kudla et al (2009,Science 10 April 2009).
Chamary and Hurst (2009) 'The price of silent mutations', Scientific
American, June 2009, pp34-41. It appears that bases in protein coding
exons can be also intron splicing recognition sites, and that a synonymous
mutation can prevent intron splicing, resulting in mutated proteins
Patterns in Evolution
I. Phylogenetic
II. Morphological
III. Historical (later)
IV. Biogeographical
Patterns in Evolution
I. Phylogenetic
- Determining the genealogical, familial patterns among organisms,
populations, species and higher taxa - "family trees"
Patterns in Evolution
I. Phylogenetic
A. Systematics: Taxonomy and Classification
Patterns in Evolution
I. Phylogenetic
A. Systematics: Taxonomy and Classification
1. Taxonomy - the naming of taxa (singular 'taxon")
Patterns in Evolution
I. Phylogenetic
A. Systematics: Taxonomy and Classification
1. Taxonomy - the naming of taxa (singular 'taxon")
a. Rules for naming species:
•Latin binomen (Drosophila melanogaster)
•italicized or underlined
•author (Drosophila melanogaster Meigen, 1830)
•if a species is named twice, priority counts
•based on a 'holotype' or 'type' specimen
•'paratypes' show range of variation
•'species' is both singular and plural; genus (s.), genera (pl.)
Patterns in Evolution
I. Phylogenetic
A. Systematics: Taxonomy and Classification
1. Taxonomy - the naming of taxa (singular 'taxon")
b. Rules for higher taxa
•Animal families end in "-idae" (Felidae)
•Animal sub-families end in "-inae" (Homininae)
•These are often derived from the same stem as the 'type genus' the first genus described for the family. (Felis)
•Plant families end in "-aceae" (Betulaceae)
•Higher taxa are capitalized, but not italicized (as above)
•adjectives are not capitalized ("hominids")
Patterns in Evolution
I. Phylogenetic
A. Systematics: Taxonomy and Classification
1. Taxonomy - the naming of taxa (singular 'taxon")
2. Classification - determining the hierarchical position of each
species within higher taxa.
A Nested Hierarchy....
Patterns in Evolution
I. Phylogenetic
A. Systematics: Taxonomy and Classification
1. Taxonomy - the naming of taxa (singular 'taxon")
2. Classification - determining the hierarchical position of each
species within higher taxa.
3. Phylogenetics
Patterns in Evolution
I. Phylogenetic
A. Systematics: Taxonomy and Classification
1. Taxonomy - the naming of taxa (singular 'taxon")
2. Classification - determining the hierarchical position of each
species within higher taxa.
3. Phylogenetics
• Cladogenesis: you want the branching/"clade" pattern of taxa to
reflect phylogenetic relationships, and you want your taxonomy to
reflect phylogenetic relationships – “Archosaurs” for
crocodilians and birds…
Patterns in Evolution
I. Phylogenetic
A. Systematics: Taxonomy and Classification
1. Taxonomy - the naming of taxa (singular 'taxon")
2. Classification - determining the hierarchical position of each
species within higher taxa.
3. Phylogenetics
• Anagenesis: however, some evolutionary changes are so
profound that we might honor the degree of difference
("Class: Aves”)
Terms:
Monophyletic taxon: includes
all (and only) the species
descended from a common
ancestor. Aves is good.
Terms:
Monophyletic taxon: includes
all (and only) the species
descended from a common
ancestor. Aves is good.
Paraphyletic taxon: includes
all descendants of a common
ancestor, except for those
placed in another taxon. So,
“Reptilia” is a paraphyletic
group, as it includes all
amniotes EXCEPT mammals
and birds (this gets the
synapsids).
c. Terms:
Monophyletic taxon: includes
all (and only) the species
descended from a common
ancestor. Aves is good.
Paraphyletic taxon: includes
all descendants of a common
ancestor, except for those
placed in another taxon. So,
“Reptilia” is a paraphyletic
group, as it includes all
diapsids and anapsids
EXCEPT mammals and birds
(this gets the synapsids).
Polyphyletic taxon: includes
organisms that do not share a
common ancestor that is in
the group. To be avoided.
“Fliers” (Birds, Pterosaurs)
Linnaean Classification of Apes
Hominidae
Pongidae
Hylobatidae
Apes = primates (grasping hands, binocular vision) with no tails
Linnaean Classification of Apes
Hylobatidae
Pongidae
PARAPHYLETIC
Linnaean Classification of Apes
Hylobatidae
Hominidae
Patterns in Evolution
I. Phylogenetic
A. Systematics: Taxonomy and Classification
B. Reconstructing Phylogenies
Patterns in Evolution
I. Phylogenetic
A. Systematics: Taxonomy and Classification
B. Reconstructing Phylogenies
1. Characters
•morphological
•behavioral
•cellular (structural or chemical)
•genetic - nitrogenous base sequence; amino acid sequence
Patterns in Evolution
I. Phylogenetic
A. Systematics: Taxonomy and Classification
B. Reconstructing Phylogenies
1. Characters
•morphological
•behavioral
•cellular (structural or chemical)
•genetic - nitrogenous base sequence; amino acid sequence
•can be quantitative measurements, or qualitative
"presence/absence“ or “A, C, T, G”
Patterns in Evolution
I. Phylogenetic
A. Systematics: Taxonomy and Classification
B. Reconstructing Phylogenies
1. Characters
2. Trees
a. Unrooted trees: show patterns among groups without specifying
ancestral relationships
Trait 1
Trait 2
Trait 3
Trait 4
Trait 5
A
0
0
0
1
1
B
0
0
1
1
1
C
1
1
1
0
1
D
1
1
0
0
1
Patterns in Evolution
I. Phylogenetic
A. Systematics: Taxonomy and Classification
B. Reconstructing Phylogenies
1. Characters
2. Trees
So, A and B share three traits that C and D don't have (1,2, 4) and are
more similar to one another than they are to C and D.
Trait 1
Trait 2
Trait 3
Trait 4
Trait 5
A
0
0
0
1
1
B
0
0
1
1
1
C
1
1
1
0
1
D
1
1
0
0
1
Patterns in Evolution
I. Phylogenetic
A. Systematics: Taxonomy and Classification
B. Reconstructing Phylogenies
1. Characters
2. Trees
Same for C and D.
Trait 1
Trait 2
Trait 3
Trait 4
Trait 5
A
0
0
0
1
1
B
0
0
1
1
1
C
1
1
1
0
1
D
1
1
0
0
1
Patterns in Evolution
I. Phylogenetic
A. Systematics: Taxonomy and Classification
B. Reconstructing Phylogenies
1. Characters
2. Trees
So, A and B share three traits that C and D don't have (1,2, 4) and
are more similar to one another than they are to C and D.
A
B
C
D
Patterns in Evolution
Trait 1
0
0
1
1
Trait 2
0
0
1
1
I. Phylogenetic
Trait 3
0
1
1
0
Trait 4
1
1
0
0
Trait 5
1
1
1
1
A. Systematics: Taxonomy and Classification
B. Reconstructing Phylogenies
1. Characters
2. Trees
b. Rooted Trees: Hypothetical patterns of descent that could be
produced with this pattern. You might suppose it would have to be
this:
A
B
C
D
Patterns in Evolution
Trait 1
0
0
1
1
Trait 2
0
0
1
1
I. Phylogenetic
Trait 3
0
1
1
0
Trait 4
1
1
0
0
Trait 5
1
1
1
1
A. Systematics: Taxonomy and Classification
B. Reconstructing Phylogenies
1. Characters
2. Trees
b. Rooted Trees: But it could easily be one of these, depending on
whether the state ‘0’ or ‘1’ for traits 1 and 2 were ancestral.
‘0’
derived
‘1’
derived
Patterns in Evolution
I. Phylogenetic
A. Systematics: Taxonomy and Classification
B. Reconstructing Phylogenies
1. Characters
2. Trees
b. Rooted Trees:
SO, in order to access ancestry, we need to compare the groups in
question to an "outgroup". An outgroup is a sister taxon which should
only share ancestral traits with the group in question. So reptiles
would be the outgroup for comparisons among diverse mammals, for
example; or a crocodile or dinosaur would be the outgroup to a
comparison among diverse birds.
Now, we assume that spE expresses ANCESTRAL characters
(plesiomorphies). Any different character state must have evolved FROM this
ancestral state - and this evolved state is called DERIVED (apomorphy).
A
B
C
D
E (out)
Trait 1
0
0
1
1
1
Trait 2
0
0
1
1
1
Trait 3
0
1
1
0
0
Trait 4
1
1
0
0
0
Trait 5
1
1
1
1
0
Now, all species in a clade might share plesiomorphies, because they are all
ultimately derived from the same ancestor. So shared ancestral traits tell us
nothing about relationships within the group. But DERIVED traits will only be
shared by species that share a more recent common ancestor...
A
B
C
D
E (out)
Trait 1
0
0
1
1
1
Trait 2
0
0
1
1
1
Trait 3
0
1
1
0
0
Trait 4
1
1
0
0
0
Trait 5
1
1
1
1
0
So, to reconstruct phylogenies and build a rooted tree, we don't just count
shared traits... we count SHARED, DERIVED traits (synapomorphies)
A
B
C
D
E (out)
Trait 1
0
0
1
1
1
Trait 2
0
0
1
1
1
Trait 3
0
1
1
0
0
Trait 4
1
1
0
0
0
Trait 5
1
1
1
1
0
So, A and B share 4 synapomorphies: 1, 2, 4, and 5 (they share these traits,
and their state is different from the outgroup). B and C share 2 synapomorphy
(3, 5).
A
B
C
D
E (out)
Trait 1
0
0
1
1
1
Trait 2
0
0
1
1
1
Trait 3
0
1
1
0
0
Trait 4
1
1
0
0
0
Trait 5
1
1
1
1
0
Number of synapomorphies:
A
B
C
B
4
-
-
C
1
2
-
D
1
1
1
Now, there are a couple rooted trees that fit these data equally well:
First, our assumed tree:
In this case, the shared trait between B and C
must be interpreted as an instance of
"convergent/parallel evolution (CE)", in which
the trait evolved independently in both
species (not inherited from ancestor).
A
4
1
1
B
C
D
B
2
1
C
1
3
1, 2,
and 4
5
A
B
C
D
E
Trait 1
0
0
1
1
1
Trait 2
0
0
1
1
1
Trait 3
0
1
1
0
0
Trait 4
1
1
0
0
0
Trait 5
1
1
1
1
0
Now, there are a couple rooted trees that fit these data equally well:
But there is another:
In this case, the discrepancy between A, B,
and C is explained as an evolutionary
"reversal" in A, which has re-expressed
the ancestral trait.
A
4
1
1
B
C
D
B
2
1
C
1
3
1, 2,
and 4
3
5
A
B
C
D
E
Trait 1
0
0
1
1
1
Trait 2
0
0
1
1
1
Trait 3
0
1
1
0
0
Trait 4
1
1
0
0
0
Trait 5
1
1
1
1
0
In both cases, species share traits for reasons OTHER than inheritance for an
immediate common ancestor. These are called homoplasies, and they
obviously can confound the reconstruction of phylogenies. Both trees require
6 evolutionary events, so they are equally "parsimonious" (simple). We could
envision lots of other trees, but they would require more reversions and
convergent events. We apply Occam's Razor - a philosophical dictum that we
will accept (and subsequently test) the simplest trees that express "maximum
parsimony". So these two trees are our phylogenetic hypotheses – to be
tested by more data that explicitly addresses their differences.
We might combine what we know from both equally parsimonius trees into a
“consensus tree”.
Both trees have clade A-B, and E as the outgroup. Relationships of C and D to
one another and to A-B is ambiguous, so a polytomy (“comb”) is used….
A
B
C
D
E
The only trait we did not define was an autapomorphy - this is a trait unique to
a species. In our examples above, each trait has only two character states. But
consider nucleotides, where each trait (position) has 4 possibilities. we can
envision that a species might have a T whereas all other species in the tree
have A, C, or G. This would be an autapomorphy, and obviously doesn't help
us out in phylogeny reconstruction because it doesn't share this trait with
anything else.