* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download LIPOchip®, a DNA-array based system
E. coli long-term evolution experiment wikipedia , lookup
Surround optical-fiber immunoassay wikipedia , lookup
Community fingerprinting wikipedia , lookup
Exome sequencing wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Personalized medicine wikipedia , lookup
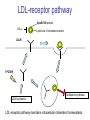
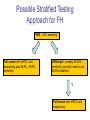
Validation of a novel mutation screening strategy for Familial Hypercholesterolaemia: LIPOchip®, a DNA-array based system Jonathan Callaway Wessex Regional Genetics Laboratory Familial Hypercholesterolaemia (FH) • Autosomal dominant disorder of lipid metabolism • Heterozygous prevalence in UK population of 1 in 500 • Characterised by: – Raised serum LDL-c (low density lipoprotein cholesterol) – Tendon and skin xanthomata (due to cholesterol deposits) – Premature coronary heart disease • Early diagnosis is beneficial to patients since treatment with lipidlowering therapy (e.g. statins) can result in a near-normal life expectancy by lowering the risk of coronary heart disease • Homozygous FH exists but is rare – Prevalence of 1 in a million – Symptoms more severe: appear in childhood and often lead to early death from coronary heart disease Genes implicated in FH • FH is a genetically heterogeneous disorder • Mutations which cosegregate with the disease have been found in at least three genes: – LDLR (low density lipoprotein receptor) • Over 1000 mutations spread throughout gene • Exonic deletions and duplications (5-10% FH cases) – APOB (apolipoprotein B) • 9 mutations – PCSK9 (proprotein convertase subtilisin/kexin type 9) • 6 mutations • Most mutations identified are in LDLR (~79%) with lower proportion in APOB (~5.5%) and PCSK9 (~1.5%) LDL-receptor pathway ApoB-100 protein LDL-c Lipid core of cholesterol esters LDLR ? PCSK9 Cholesterol synthesis LDLR synthesis LDL-receptor pathway maintains intracellular cholesterol homeostasis Current FH testing strategy • FH20 Elucigene ARMS kit (Tepnel Diagnostics) – Identifies 20 most common mutations in UK population – Sensitivity of only 40% • NICE guidelines recommend DNA testing be used to confirm a diagnosis of FH (March 2009) • Need for an increase in testing sensitivity LIPOchip® • A DNA array-based system designed by Progenika • Used as the primary testing strategy for FH in Spain • Detection of 251 common FH point mutations – 242 LDLR, 3 APOB, 6 PCSK9 • Copy number variation detection in LDLR • Currently targeted towards Spanish population within which the manufacturers claim a sensitivity of 80% • 4 FH20 mutations are not detected by Spanish version – FS206, K290RfsX20, Q363X, C656R • British version is under development – June 2010 availability – Sensitivity of 80-85% – Probe sets for the 4 missing FH20 mutations LIPOchip®: DNA-array technology 1. 2. 3. 4. Multiplex PCR amplification Product fragmentation Labelling with biotin Hybridization and washing - Using Tecan 4800 HS Pro station Addition of Cy3-streptavidin (fluorochrome) 5. Results analysis - Using Agilent scanner and customised software Light Laser Cy3-streptavidin Biotin Fragmented PCR product A scanned LIPOchip® slide • 2 pairs of oligonucleotides per mutation: – Each pair consists of a WT probe and a mutant probe – Signal intensity ratios calculated for WT / (WT+Mut) • Controls for hybridization process and for measuring background signal noise • Copy number variation detection controls Graphical display of results generated by LIPOchip® software Normal Heterozygous WT/mutant Homozygous mutant Validation Strategy • 48 LIPOchip® slides were provided by Progenika to validate the technology • Maximum of 12 samples per run - in order to perform copy number detection two of these must be normal male & female controls • Samples selected for validation: – – – • 10 normal controls 6 FH20 positive controls 22 FH20 negative patients Criteria for selection of FH20 negative patients: – – ‘Definite FH’ on referral card; or High cholesterol level (over 8 mmol/L) plus either (i) Family history of high cholesterol; or (ii) Family history of cardiovascular disease Results from Validation • Normal controls: – 9/10 slides passed quality control – No point mutations were detected • FH20 positive controls: – 6/6 mutations correctly called by LIPOchip® • FH20 negative patients: – 2 pathogenic LDLR missense mutations: • c.1796T>C (p.Leu599Ser) • c.1618G>A (p.Ala540Thr) – 1 unclassified LDLR variant: • c.2177C>T (p.Thr726Ile)…likely non-pathogenic by in-silico analysis – Mutations were confirmed by direct sequencing Problem 1: The M064 probe set Normal • c.91G>T (p.Glu31X) • ‘No Call’ result was frequently obtained: – 7/9 normal controls – 3/6 positive controls – 20/22 FH20 negative patients • Signal intensity values extended beyond the normal distribution parameters although they were still distinct from the mutation range • Progenika are aware of this problem and hope to resolve it in the forthcoming British version Problem 2: Copy number variation detection 7/9 normal controls appeared to have a deletion of the LDLR promoter and exon 1 Normal LDLR gene dosage Apparent deletion of promoter + exon 1 Problem 2: Copy number variation detection Also, poor quality dosage data was often generated… These issues raised the question as to whether LIPOchip® could be used in our laboratory for reliable copy number variation detection Traditional Full Screen • Testing strategy – Combination of dHPLC and direct sequencing of LDLR gene – MLPA for dosage analysis of LDLR gene (MRC-Holland kit P062-C1) • Samples: – 10 normals from the validation – 22 FH20 negative patients tested using LIPOchip® Results from Traditional Full Screen • The 2 pathogenic LDLR mutations and the unclassified LDLR variant identified by LIPOchip® were confirmed • 2 further pathogenic LDLR mutations and an additional unclassified variant were detected: – c.1061A>T (p.Asp354Val) – c.1067delA (p.Ala356ValfsX14) – c.2479G>A (p.Val827Ile)…undecided pathogenicity by in-silico analysis • MLPA did not detect any deletions or duplications in the LDLR promoter or exonic regions of patients or controls Possible Stratified Testing Approach for FH FH20 – 40% sensitivity Full screen with dHPLC and sequencing plus MLPA – 99.9% sensitivity LIPOchip® - possibly 80-85% sensitivity (currently need to use MLPA in addition) ? Full screen with dHPLC and sequencing Implementation of LIPOchip®? Full screen LIPOchip® Sensitivity : Cost Ratio Conclusions • LIPOchip® can be reliably used to detect common FH point mutations with an increase in testing sensitivity • Currently MLPA is required as a necessary complement to LIPOchip® testing • Some mutations detected by LIPOchip® require further investigation regarding their pathogenicity using in-silico analysis • Additional validation work is needed on the British version of LIPOchip®, when available • Costing is an issue for LIPOchip® and will influence the decision on whether or not to use the technology in a diagnostic setting Acknowledgements • Wessex Regional Genetics Laboratory: – Oliver Wood – Esta Cross – Alison Skinner – Dr John Harvey • Progenika: – Dr Xabier Abad Lloret – Maximiliano Crosetti