* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Genetic dissection of trisomy 21 pathology using a
Genetic engineering wikipedia , lookup
Y chromosome wikipedia , lookup
Genomic imprinting wikipedia , lookup
Medical genetics wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Designer baby wikipedia , lookup
History of genetic engineering wikipedia , lookup
Microevolution wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
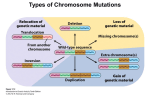
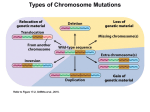
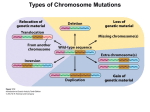
Down syndrome wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Neocentromere wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Genetic dissection of trisomy 21 pathology using a transchromosomic mouse Down Syndrome model. C. Canzonetta, S. Devita, E.M Fisher, V. Tybulewicz, J. Groet and Dean Nizetic Since Down Syndrome (DS) is not an inherited disease, and the DNA sequence of the supernumerary chromosome 21 causing it is perfectly normal, standard genetics cannot be used to pinpoint the causative roles of individual genes in the disorder-component symptoms of DS. Studies of partial trisomies have helped to delimit the chromosomal segments associated with clinical phenotypes. Taking advantage of a transchromosomic Down Syndrome mouse model (mouse cells bearing supernumerary human chromosome 21) we developed a novel genetic dissection system. Transchromosomic embryonic stem cells (ESC) were successfully used to define cellular phenotypes reproducing in primary cells from foeti with trisomy 21 (T21), and in foetal T21 tissues. Using a combination of partial transchromosomic cell lines, and an approach of specific correction-to-disomy of one candidate gene at a time (while maintaining the remainder of the chromosome in trisomy) it has been possible to pinpoint genes, whose correction to disomy removes the cellular phenotype observed. Using the transchromosomic ESC, DYRK1A was found responsible for causing the earliest hitherto reported disturbance in the molecular make-up of T21, the de-regulation of the NRSF/REST regulon, the master controller of neuronal differentiation. A major disturbance of the transcriptional circuitry regulating ESC pluripotency and lineage determination was also observed. The earliest stages of haematopoietic commitment (mesodermal colony formation) were also analised in vitro leading to the obsevation that trisomy 21 causes increased production of haemogenic endothelial cells, haematopoietic stem cell (HSC) precursors, and increased colony forming potential, with significantly increased immature progenitors. The use of human-specific siRNA silencing, lead to the demonstration that an extra copy of RUNX1 is at the basis of the imbalance which in ultimate analysis can be linked to Down Syndrome individuals propensity to develop various types of leukemia.